Improving natural language information extraction from cancer pathology reports using transfer learning and zero-shot string similarity
- PMID: 34604711
- PMCID: PMC8484934
- DOI: 10.1093/jamiaopen/ooab085
Improving natural language information extraction from cancer pathology reports using transfer learning and zero-shot string similarity
Abstract
Objective: We develop natural language processing (NLP) methods capable of accurately classifying tumor attributes from pathology reports given minimal labeled examples. Our hierarchical cancer to cancer transfer (HCTC) and zero-shot string similarity (ZSS) methods are designed to exploit shared information between cancers and auxiliary class features, respectively, to boost performance using enriched annotations which give both location-based information and document level labels for each pathology report.
Materials and methods: Our data consists of 250 pathology reports each for kidney, colon, and lung cancer from 2002 to 2019 from a single institution (UCSF). For each report, we classified 5 attributes: procedure, tumor location, histology, grade, and presence of lymphovascular invasion. We develop novel NLP techniques involving transfer learning and string similarity trained on enriched annotations. We compare HCTC and ZSS methods to the state-of-the-art including conventional machine learning methods as well as deep learning methods.
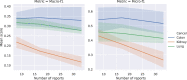
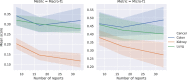
Results: For our HCTC method, we see an improvement of up to 0.1 micro-F1 score and 0.04 macro-F1 averaged across cancer and applicable attributes. For our ZSS method, we see an improvement of up to 0.26 micro-F1 and 0.23 macro-F1 averaged across cancer and applicable attributes. These comparisons are made after adjusting training data sizes to correct for the 20% increase in annotation time for enriched annotations compared to ordinary annotations.
Conclusions: Methods based on transfer learning across cancers and augmenting information methods with string similarity priors can significantly reduce the amount of labeled data needed for accurate information extraction from pathology reports.
Keywords: cancer; natural language processing; pathology.
© The Author(s) 2021. Published by Oxford University Press on behalf of the American Medical Informatics Association.
Figures
Similar articles
-
Supervised line attention for tumor attribute classification from pathology reports: Higher performance with less data.J Biomed Inform. 2021 Oct;122:103872. doi: 10.1016/j.jbi.2021.103872. Epub 2021 Aug 16. J Biomed Inform. 2021. PMID: 34411709
-
Privacy-Preserving Deep Learning NLP Models for Cancer Registries.IEEE Trans Emerg Top Comput. 2021 Jul-Sep;9(3):1219-1230. doi: 10.1109/tetc.2020.2983404. Epub 2020 Apr 16. IEEE Trans Emerg Top Comput. 2021. PMID: 36117774 Free PMC article.
-
Automatic extraction of cancer registry reportable information from free-text pathology reports using multitask convolutional neural networks.J Am Med Inform Assoc. 2020 Jan 1;27(1):89-98. doi: 10.1093/jamia/ocz153. J Am Med Inform Assoc. 2020. PMID: 31710668 Free PMC article.
-
A Question-and-Answer System to Extract Data From Free-Text Oncological Pathology Reports (CancerBERT Network): Development Study.J Med Internet Res. 2022 Mar 23;24(3):e27210. doi: 10.2196/27210. J Med Internet Res. 2022. PMID: 35319481 Free PMC article.
-
Natural language processing in clinical neuroscience and psychiatry: A review.Front Psychiatry. 2022 Sep 14;13:946387. doi: 10.3389/fpsyt.2022.946387. eCollection 2022. Front Psychiatry. 2022. PMID: 36186874 Free PMC article.
Cited by
-
TCGA-Reports: A machine-readable pathology report resource for benchmarking text-based AI models.Patterns (N Y). 2024 Feb 21;5(3):100933. doi: 10.1016/j.patter.2024.100933. eCollection 2024 Mar 8. Patterns (N Y). 2024. PMID: 38487800 Free PMC article.
References
-
- Burger G, Abu-Hanna A, de Keizer N, et al.Natural language processing in pathology: a scoping review. J Clin Pathol 2016; jclinpath-2016-203872. - PubMed
-
- Martinez D, Yue L. Information extraction from pathology reports in a hospital setting. In: Proceedings of the 20th ACM International Conference on Information and Knowledge Management. 2011: 1877–82; Glasgow Scotland, UK.
-
- Napolitano G, Fox C, Middleton R, et al.Pattern-based information extraction from pathology reports for cancer registration. Cancer Causes Control 2010; 21 (11): 1887–94. - PubMed
