UnCovid: A versatile, low-cost, and open-source protocol for SARS-CoV-2 RNA detection
- PMID: 34604812
- PMCID: PMC8463329
- DOI: 10.1016/j.xpro.2021.100878
UnCovid: A versatile, low-cost, and open-source protocol for SARS-CoV-2 RNA detection
Abstract
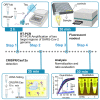
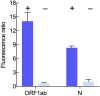
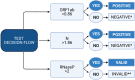
Here, we describe a detailed step-by-step protocol to detect SARS-CoV-2 RNA using RT-PCR-mediated amplification and CRISPR/Cas-based visualization. The optimized assay uses basic molecular biology equipment such as conventional thermocyclers and transilluminators for qualitative detection. Alternatively, a fluorescence plate reader can be used for quantitative measurements. The protocol detects two regions of the SARS-CoV-2 genome in addition to the human RNaseP sample control. Aiming to reach remote regions, this work was developed to use the portable molecular workstation from BentoLab. For complete details on the use and execution of this protocol, please refer to Alcántara et al., 2021.
Keywords: Biotechnology and bioengineering; CRISPR; Clinical Protocol; Immunology; Microbiology; Molecular Biology.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Comment in
-
Open access methods and protocols promote open science in a pandemic.STAR Protoc. 2022 Mar 2;3(1):101226. doi: 10.1016/j.xpro.2022.101226. eCollection 2022 Mar 18. STAR Protoc. 2022. PMID: 35284832 Free PMC article.
References
-
- Álvarez-Fernández R. Chapter one explanatory chapter PCR primer design. Methods Enzymol. 2013;529:1–21. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

