Metalloprotein catalysis: structural and mechanistic insights into oxidoreductases from neutron protein crystallography
- PMID: 34605429
- PMCID: PMC8489226
- DOI: 10.1107/S2059798321009025
Metalloprotein catalysis: structural and mechanistic insights into oxidoreductases from neutron protein crystallography
Abstract
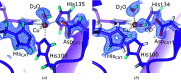
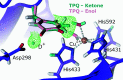
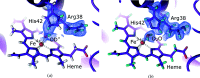
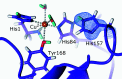
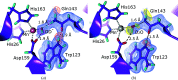
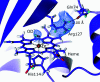
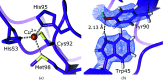
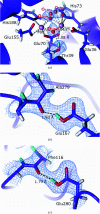
Metalloproteins catalyze a range of reactions, with enhanced chemical functionality due to their metal cofactor. The reaction mechanisms of metalloproteins have been experimentally characterized by spectroscopy, macromolecular crystallography and cryo-electron microscopy. An important caveat in structural studies of metalloproteins remains the artefacts that can be introduced by radiation damage. Photoreduction, radiolysis and ionization deriving from the electromagnetic beam used to probe the structure complicate structural and mechanistic interpretation. Neutron protein diffraction remains the only structural probe that leaves protein samples devoid of radiation damage, even when data are collected at room temperature. Additionally, neutron protein crystallography provides information on the positions of light atoms such as hydrogen and deuterium, allowing the characterization of protonation states and hydrogen-bonding networks. Neutron protein crystallography has further been used in conjunction with experimental and computational techniques to gain insight into the structures and reaction mechanisms of several transition-state metal oxidoreductases with iron, copper and manganese cofactors. Here, the contribution of neutron protein crystallography towards elucidating the reaction mechanism of metalloproteins is reviewed.
Keywords: X-ray diffraction; enzymatic mechanisms; metalloproteins; neutron protein crystallography; protonation; radiation damage.
open access.
Figures

Similar articles
-
Combining X-ray and neutron crystallography with spectroscopy.Acta Crystallogr D Struct Biol. 2017 Feb 1;73(Pt 2):141-147. doi: 10.1107/S2059798316016314. Epub 2017 Feb 1. Acta Crystallogr D Struct Biol. 2017. PMID: 28177310 Free PMC article.
-
Neutron Crystallography Data Collection and Processing for Modelling Hydrogen Atoms in Protein Structures.J Vis Exp. 2020 Dec 1;(166). doi: 10.3791/61903. J Vis Exp. 2020. PMID: 33346193
-
Neutron crystallographic refinement with REFMAC5 from the CCP4 suite.Acta Crystallogr D Struct Biol. 2023 Dec 1;79(Pt 12):1056-1070. doi: 10.1107/S2059798323008793. Epub 2023 Nov 3. Acta Crystallogr D Struct Biol. 2023. PMID: 37921806 Free PMC article.
-
Sub-atomic resolution X-ray crystallography and neutron crystallography: promise, challenges and potential.IUCrJ. 2015 Jun 30;2(Pt 4):464-74. doi: 10.1107/S2052252515011239. eCollection 2015 Jul 1. IUCrJ. 2015. PMID: 26175905 Free PMC article. Review.
-
Neutron protein crystallography: A complementary tool for locating hydrogens in proteins.Arch Biochem Biophys. 2016 Jul 15;602:48-60. doi: 10.1016/j.abb.2015.11.033. Epub 2015 Nov 22. Arch Biochem Biophys. 2016. PMID: 26592456 Review.
Cited by
-
Perdeuterated GbpA Enables Neutron Scattering Experiments of a Lytic Polysaccharide Monooxygenase.ACS Omega. 2023 Jul 31;8(32):29101-29112. doi: 10.1021/acsomega.3c02168. eCollection 2023 Aug 15. ACS Omega. 2023. PMID: 37599915 Free PMC article.
-
Joint X-ray/neutron structure of Lentinus similis AA9_A at room temperature.Acta Crystallogr F Struct Biol Commun. 2023 Jan 1;79(Pt 1):1-7. doi: 10.1107/S2053230X22011335. Epub 2023 Jan 1. Acta Crystallogr F Struct Biol Commun. 2023. PMID: 36598350 Free PMC article.
-
The Biochemistry and Effectiveness of Antioxidants in Food, Fruits, and Marine Algae.Antioxidants (Basel). 2023 Apr 2;12(4):860. doi: 10.3390/antiox12040860. Antioxidants (Basel). 2023. PMID: 37107235 Free PMC article. Review.
-
Quantum refinement in real and reciprocal space using the Phenix and ORCA software.IUCrJ. 2024 Nov 1;11(Pt 6):921-937. doi: 10.1107/S2052252524008406. IUCrJ. 2024. PMID: 39345101 Free PMC article.
-
Exploring the World of Membrane Proteins: Techniques and Methods for Understanding Structure, Function, and Dynamics.Molecules. 2023 Oct 19;28(20):7176. doi: 10.3390/molecules28207176. Molecules. 2023. PMID: 37894653 Free PMC article. Review.
References
-
- Abreu, I. A. & Cabelli, D. E. (2010). Biochim. Biophys. Acta, 1804, 263–274. - PubMed
-
- Andreini, C., Bertini, I., Cavallaro, G., Holliday, G. L. & Thornton, J. M. (2008). J. Biol. Inorg. Chem. 13, 1205–1218. - PubMed
-
- Antonyuk, S. V., Melik-Adamyan, V. R., Popov, A. N., Lamzin, V. S., Hempstead, P. D., Harrison, P. M., Artymyuk, P. J. & Barynin, V. V. (2000). Crystallogr. Rep. 45, 105–116.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

