Determination of intracellular protein-ligand binding affinity by competition binding in-cell NMR
- PMID: 34605430
- PMCID: PMC8489230
- DOI: 10.1107/S2059798321009037
Determination of intracellular protein-ligand binding affinity by competition binding in-cell NMR
Abstract
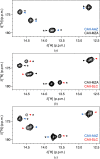
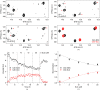
Structure-based drug development suffers from high attrition rates due to the poor activity of lead compounds in cellular and animal models caused by low cell penetrance, off-target binding or changes in the conformation of the target protein in the cellular environment. The latter two effects cause a change in the apparent binding affinity of the compound, which is indirectly assessed by cellular activity assays. To date, direct measurement of the intracellular binding affinity remains a challenging task. In this work, in-cell NMR spectroscopy was applied to measure intracellular dissociation constants in the nanomolar range by means of protein-observed competition binding experiments. Competition binding curves relative to a reference compound could be retrieved either from a series of independent cell samples or from a single real-time NMR bioreactor run. The method was validated using a set of sulfonamide-based inhibitors of human carbonic anhydrase II with known activity in the subnanomolar to submicromolar range. The intracellular affinities were similar to those obtained in vitro, indicating that these compounds selectively bind to the intracellular target. In principle, the approach can be applied to any soluble intracellular target that gives rise to measurable chemical shift changes upon ligand binding.
Keywords: bioreactor; carbonic anhydrase inhibitors; competition binding; in-cell NMR; real-time NMR.
open access.
Figures




Similar articles
-
Carbonic anhydrase inhibitors: Synthesis and inhibition of the cytosolic mammalian carbonic anhydrase isoforms I, II and VII with benzene sulfonamides incorporating 4,5,6,7-tetrachlorophthalimide moiety.Bioorg Med Chem. 2013 Sep 1;21(17):5168-74. doi: 10.1016/j.bmc.2013.06.035. Epub 2013 Jun 26. Bioorg Med Chem. 2013. PMID: 23867389
-
Ureidobenzenesulfonamides as efficient inhibitors of carbonic anhydrase II.Bioorg Chem. 2019 Oct;91:103123. doi: 10.1016/j.bioorg.2019.103123. Epub 2019 Jul 17. Bioorg Chem. 2019. PMID: 31336306
-
Highly hydrophilic 1,3-oxazol-5-yl benzenesulfonamide inhibitors of carbonic anhydrase II for reduction of glaucoma-related intraocular pressure.Bioorg Med Chem. 2019 Nov 1;27(21):115086. doi: 10.1016/j.bmc.2019.115086. Epub 2019 Sep 4. Bioorg Med Chem. 2019. PMID: 31515057
-
Intracellular Binding/Unbinding Kinetics of Approved Drugs to Carbonic Anhydrase II Observed by in-Cell NMR.ACS Chem Biol. 2020 Oct 16;15(10):2792-2800. doi: 10.1021/acschembio.0c00590. Epub 2020 Sep 30. ACS Chem Biol. 2020. PMID: 32955851 Free PMC article.
-
Affinity measurement of strong ligands with NMR spectroscopy: Limitations and ways to overcome them.Prog Nucl Magn Reson Spectrosc. 2023 Nov-Dec;138-139:52-69. doi: 10.1016/j.pnmrs.2023.07.001. Epub 2023 Jul 18. Prog Nucl Magn Reson Spectrosc. 2023. PMID: 38065668 Review.
Cited by
-
Ligand-Based Competition Binding by Real-Time 19F NMR in Human Cells.J Med Chem. 2024 Jan 25;67(2):1115-1126. doi: 10.1021/acs.jmedchem.3c01600. Epub 2024 Jan 12. J Med Chem. 2024. PMID: 38215028 Free PMC article.
-
Intracellular drug binding affinities by NMR.Acta Crystallogr D Struct Biol. 2021 Oct 1;77(Pt 10):1216-1217. doi: 10.1107/S2059798321010135. Epub 2021 Oct 1. Acta Crystallogr D Struct Biol. 2021. PMID: 34605425 Free PMC article.
-
Radio Signals from Live Cells: The Coming of Age of In-Cell Solution NMR.Chem Rev. 2022 May 25;122(10):9267-9306. doi: 10.1021/acs.chemrev.1c00790. Epub 2022 Jan 21. Chem Rev. 2022. PMID: 35061391 Free PMC article. Review.
-
Direct Expression of Fluorinated Proteins in Human Cells for 19F In-Cell NMR Spectroscopy.J Am Chem Soc. 2023 Jan 18;145(2):1389-1399. doi: 10.1021/jacs.2c12086. Epub 2023 Jan 5. J Am Chem Soc. 2023. PMID: 36604341 Free PMC article.
-
Perspectives on Applications of 19F-NMR in Fragment-Based Drug Discovery.Molecules. 2024 Dec 5;29(23):5748. doi: 10.3390/molecules29235748. Molecules. 2024. PMID: 39683906 Free PMC article. Review.
References
-
- Aricescu, A. R., Lu, W. & Jones, E. Y. (2006). Acta Cryst. D62, 1243–1250. - PubMed
-
- Banerjee, A. L., Swanson, M., Mallik, S. & Srivastava, D. K. (2004). Protein Expr. Purif. 37, 450–454. - PubMed
-
- Barbieri, L. & Luchinat, E. (2021). J. Vis. Exp., e62323. - PubMed
-
- Barbieri, L., Luchinat, E. & Banci, L. (2016). Nat. Protoc. 11, 1101–1111. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

