Structural characterization of human peptidyl-arginine deiminase type III by X-ray crystallography
- PMID: 34605437
- PMCID: PMC8488854
- DOI: 10.1107/S2053230X21009195
Structural characterization of human peptidyl-arginine deiminase type III by X-ray crystallography
Abstract
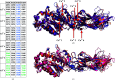
The Ca2+-dependent enzyme peptidyl-arginine deiminase type III (PAD3) catalyses the deimination of arginine residues to form citrulline residues in proteins such as keratin, filaggrin and trichohyalin. This is an important post-translation modification that is required for normal hair and skin formation in follicles and keratocytes. The structure of apo human PAD3 was determined by X-ray crystallography to a resolution of 2.8 Å. The structure of PAD3 revealed a similar overall architecture to other PAD isoforms: the N-terminal and middle domains of PAD3 show sequence and structural variety, whereas the sequence and structure of the C-terminal catalytic domain is highly conserved. Structural analysis indicates that PAD3 is a dimer in solution, as is also the case for the PAD2 and PAD4 isoforms but not the PAD1 isoform.
Keywords: calcium binding; hair follicles; peptidyl-arginine deiminase; post-translational modifications; protein citrullination.
Figures


References
-
- Arita, K., Hashimoto, H., Shimizu, T., Nakashima, K., Yamada, M. & Sato, M. (2004). Nat. Struct. Mol. Biol. 11, 777–783. - PubMed
-
- Assohou-Luty, C., Raijmakers, R., Benckhuijsen, W. E., Stammen-Vogelzangs, J., de Ru, A., van Veelen, P. A., Franken, K. L., Drijfhout, J. W. & Pruijn, G. J. (2014). Biochim. Biophys. Acta, 1844, 829–836. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

