PeptiCHIP: A Microfluidic Platform for Tumor Antigen Landscape Identification
- PMID: 34605646
- PMCID: PMC8552492
- DOI: 10.1021/acsnano.1c04371
PeptiCHIP: A Microfluidic Platform for Tumor Antigen Landscape Identification
Abstract
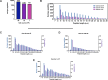
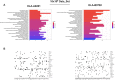
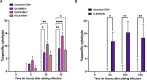
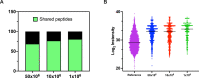
Identification of HLA class I ligands from the tumor surface (ligandome or immunopeptidome) is essential for designing T-cell mediated cancer therapeutic approaches. However, the sensitivity of the process for isolating MHC-I restricted tumor-specific peptides has been the major limiting factor for reliable tumor antigen characterization, making clear the need for technical improvement. Here, we describe our work from the fabrication and development of a microfluidic-based chip (PeptiCHIP) and its use to identify and characterize tumor-specific ligands on clinically relevant human samples. Specifically, we assessed the potential of immobilizing a pan-HLA antibody on solid surfaces via well-characterized streptavidin-biotin chemistry, overcoming the limitations of the cross-linking chemistry used to prepare the affinity matrix with the desired antibodies in the immunopeptidomics workflow. Furthermore, to address the restrictions related to the handling and the limited availability of tumor samples, we further developed the concept toward the implementation of a microfluidic through-flow system. Thus, the biotinylated pan-HLA antibody was immobilized on streptavidin-functionalized surfaces, and immune-affinity purification (IP) was carried out on customized microfluidic pillar arrays made of thiol-ene polymer. Compared to the standard methods reported in the field, our methodology reduces the amount of antibody and the time required for peptide isolation. In this work, we carefully examined the specificity and robustness of our customized technology for immunopeptidomics workflows. We tested this platform by immunopurifying HLA-I complexes from 1 × 106 cells both in a widely studied B-cell line and in patients-derived ex vivo cell cultures, instead of 5 × 108 cells as required in the current technology. After the final elution in mild acid, HLA-I-presented peptides were identified by tandem mass spectrometry and further investigated by in vitro methods. These results highlight the potential to exploit microfluidics-based strategies in immunopeptidomics platforms and in personalized immunopeptidome analysis from cells isolated from individual tumor biopsies to design tailored cancer therapeutic vaccines. Moreover, the possibility to integrate multiple identical units on a single chip further improves the throughput and multiplexing of these assays with a view to clinical needs.
Keywords: HLA peptides; affinity purification; ligandome; microfluidics; thiol−enes.
Conflict of interest statement
The authors declare the following competing financial interest(s): Vincenzo Cerullo is a cofounder and shareholder at VALO Therapeutics. The other authors have no conflicts of interest.
Figures



References
-
- Bassani-Sternberg M.; Pletscher-Frankild S.; Jensen L. J.; Mann M. Mass Spectrometry of Human Leukocyte Antigen Class I Peptidomes Reveals Strong Effects of Protein Abundance and Turnover on Antigen Presentation. Mol. Cell Proteomics. 2015, 14 (3), 658–73. 10.1074/mcp.M114.042812. - DOI - PMC - PubMed
-
- Dutoit V.; Herold-Mende C.; Hilf N.; Schoor O.; Beckhove P.; Bucher J.; Dorsch K.; Flohr S.; Fritsche J.; Lewandrowski P.; Lohr J.; Rammensee H. G.; Stevanovic S.; Trautwein C.; Vass V.; Walter S.; Walker P. R.; Weinschenk T.; Singh-Jasuja H.; Dietrich P. Y. Exploiting the Glioblastoma Peptidome to Discover Novel Tumour-Associated Antigens for Immunotherapy. Brain 2012, 135 (4), 1042–1054. 10.1093/brain/aws042. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

