Identification and validation of potential novel biomarkers for oral squamous cell carcinoma
- PMID: 34606406
- PMCID: PMC8806987
- DOI: 10.1080/21655979.2021.1987089
Identification and validation of potential novel biomarkers for oral squamous cell carcinoma
Abstract
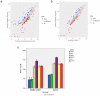
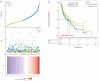
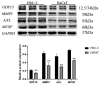
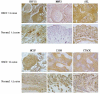
Our study aimed to explore potential new diagnostic biomarkers in patients with oral squamous cell carcinoma (OSCC) to find new target molecules involved in the progression of OSCC. Potential novel biomarkers of OSCC were identified using a protein microarray assay. Compared with the healthy control group, there were five proteins (I309, GDF15, AXL, MMP3, and CTACK) in the serum of in situ oral cancer group. However, there were four differentially expressed proteins (MCSF, I309, MMP3, and CTACK) in the serum of the OSCC group. Receiver operating characteristic (ROC) curve analysis results suggested that these six proteins (I309, GDF15, AXL, MMP3, CTACK, and MCSF) had diagnostic value for OSCC. Based on The Cancer Genome Atlas (TCGA) database, we found that only GDF15 expression was associated with the prognosis of OSCC. Subsequently, we verified the expression levels of six proteins in HSC-3 and HaCaT cells, and the results showed that the level of these six proteins was significantly higher in HSC-3 cells than in normal HaCaT cells. Similarly, in the OSCC nude mouse model, the expression levels of these proteins were significantly upregulated in OSCC tumor tissue compared to the normal tissue. GDF15, MMP3, AXL, MCSF, I309, and CTACK may be used as biomarkers for OSCC diagnosis and provide a novel study direction for the treatment of OSCC.
Keywords: Oral squamous cell carcinoma; biomarkers; diagnosis; protein microarray assay; serum.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures






References
-
- Feng X, Luo Q, Wang H, et al. MicroRNA-22 suppresses cell proliferation, migration and invasion in oral squamous cell carcinoma by targeting NLRP3. J Cell Physiol. 2018;233(9):6705–6713. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
