Scope2Screen: Focus+Context Techniques for Pathology Tumor Assessment in Multivariate Image Data
- PMID: 34606456
- PMCID: PMC8805697
- DOI: 10.1109/TVCG.2021.3114786
Scope2Screen: Focus+Context Techniques for Pathology Tumor Assessment in Multivariate Image Data
Abstract
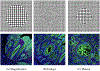
Inspection of tissues using a light microscope is the primary method of diagnosing many diseases, notably cancer. Highly multiplexed tissue imaging builds on this foundation, enabling the collection of up to 60 channels of molecular information plus cell and tissue morphology using antibody staining. This provides unique insight into disease biology and promises to help with the design of patient-specific therapies. However, a substantial gap remains with respect to visualizing the resulting multivariate image data and effectively supporting pathology workflows in digital environments on screen. We, therefore, developed Scope2Screen, a scalable software system for focus+context exploration and annotation of whole-slide, high-plex, tissue images. Our approach scales to analyzing 100GB images of 109 or more pixels per channel, containing millions of individual cells. A multidisciplinary team of visualization experts, microscopists, and pathologists identified key image exploration and annotation tasks involving finding, magnifying, quantifying, and organizing regions of interest (ROIs) in an intuitive and cohesive manner. Building on a scope-to-screen metaphor, we present interactive lensing techniques that operate at single-cell and tissue levels. Lenses are equipped with task-specific functionality and descriptive statistics, making it possible to analyze image features, cell types, and spatial arrangements (neighborhoods) across image channels and scales. A fast sliding-window search guides users to regions similar to those under the lens; these regions can be analyzed and considered either separately or as part of a larger image collection. A novel snapshot method enables linked lens configurations and image statistics to be saved, restored, and shared with these regions. We validate our designs with domain experts and apply Scope2Screen in two case studies involving lung and colorectal cancers to discover cancer-relevant image features.
Figures
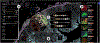



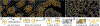
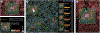


References
-
- Lensing, an npm package, https://www.npmjs.com/package/lensing, last accessed: 8/06/2021.
-
- The OME-TIFF format — OME Data Model and File Formats 5.6.3 documentation -https://docs.openmicroscopy.org/ome-model/5.6.3/ome-tiff/, last accessed: 3/31/2021.
-
- OpenSeadragon - An open-source, web-based viewer for high-resolution zoomable images - https://openseadragon.github.io, last accessed: 3/31/2021.
-
- Scope2screen codebase, https://github.com/labsyspharm/scope2screen, last accessed: 8/06/2021.
-
- Sliding window histogram, skimage: image processing in python, v0.18.0 docs - scikit-image.org/docs/stable/auto_examples/features_detection/plot_windo..., last accessed 3/31/2021.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

