Identification and classification of antiviral defence systems in bacteria and archaea with PADLOC reveals new system types
- PMID: 34606606
- PMCID: PMC8565338
- DOI: 10.1093/nar/gkab883
Identification and classification of antiviral defence systems in bacteria and archaea with PADLOC reveals new system types
Abstract
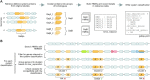
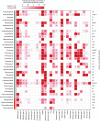
To provide protection against viral infection and limit the uptake of mobile genetic elements, bacteria and archaea have evolved many diverse defence systems. The discovery and application of CRISPR-Cas adaptive immune systems has spurred recent interest in the identification and classification of new types of defence systems. Many new defence systems have recently been reported but there is a lack of accessible tools available to identify homologs of these systems in different genomes. Here, we report the Prokaryotic Antiviral Defence LOCator (PADLOC), a flexible and scalable open-source tool for defence system identification. With PADLOC, defence system genes are identified using HMM-based homologue searches, followed by validation of system completeness using gene presence/absence and synteny criteria specified by customisable system classifications. We show that PADLOC identifies defence systems with high accuracy and sensitivity. Our modular approach to organising the HMMs and system classifications allows additional defence systems to be easily integrated into the PADLOC database. To demonstrate application of PADLOC to biological questions, we used PADLOC to identify six new subtypes of known defence systems and a putative novel defence system comprised of a helicase, methylase and ATPase. PADLOC is available as a standalone package (https://github.com/padlocbio/padloc) and as a webserver (https://padloc.otago.ac.nz).
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

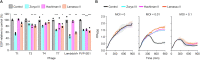
References
-
- Hampton H.G., Watson B.N.J., Fineran P.C.. The arms race between bacteria and their phage foes. Nature. 2020; 577:327–336. - PubMed
-
- Samson J.E., Magadán A.H., Sabri M., Moineau S.. Revenge of the phages: defeating bacterial defences. Nat. Rev. Microbiol. 2013; 11:675–687. - PubMed
-
- Anzalone A.V., Koblan L.W., Liu D.R.. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020; 38:824–844. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

