Efficient and high-throughput pseudorecombinant-chimeric Cucumber mosaic virus-based VIGS in maize
- PMID: 34606612
- PMCID: PMC8644855
- DOI: 10.1093/plphys/kiab443
Efficient and high-throughput pseudorecombinant-chimeric Cucumber mosaic virus-based VIGS in maize
Abstract
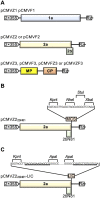
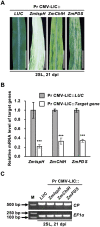
Virus-induced gene silencing (VIGS) is a versatile and attractive approach for functional gene characterization in plants. Although several VIGS vectors for maize (Zea mays) have been previously developed, their utilities are limited due to low viral infection efficiency, insert instability, short maintenance of silencing, inadequate inoculation method, or abnormal requirement of growth temperature. Here, we established a Cucumber mosaic virus (CMV)-based VIGS system for efficient maize gene silencing that overcomes many limitations of VIGS currently available for maize. Using two distinct strains, CMV-ZMBJ and CMV-Fny, we generated a pseudorecombinant-chimeric (Pr) CMV. Pr CMV showed high infection efficacy but mild viral symptoms in maize. We then constructed Pr CMV-based vectors for VIGS, dubbed Pr CMV VIGS. Pr CMV VIGS is simply performed by mechanical inoculation of young maize leaves with saps of Pr CMV-infected Nicotiana benthamiana under normal growth conditions. Indeed, suppression of isopentenyl/dimethylallyl diphosphate synthase (ZmIspH) expression by Pr CMV VIGS resulted in non-inoculated leaf bleaching as early as 5 d post-inoculation (dpi) and exhibited constant and efficient systemic silencing over the whole maize growth period up to 105 dpi. Furthermore, utilizing a ligation-independent cloning (LIC) strategy, we developed a modified Pr CMV-LIC VIGS vector, allowing easy gene cloning for high-throughput silencing in maize. Thus, our Pr CMV VIGS system provides a much-improved toolbox to facilitate efficient and long-duration gene silencing for large-scale functional genomics in maize, and our pseudorecombination-chimera combination strategy provides an approach to construct efficient VIGS systems in plants.
© American Society of Plant Biologists 2021. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures



Similar articles
-
An efficient virus-induced gene silencing vector for maize functional genomics research.Plant J. 2016 Apr;86(1):102-15. doi: 10.1111/tpj.13142. Plant J. 2016. PMID: 26921244
-
Use of a Virus Gene Silencing Vector for Maize Functional Genomics Research.Methods Mol Biol. 2018;1676:141-150. doi: 10.1007/978-1-4939-7315-6_8. Methods Mol Biol. 2018. PMID: 28986908
-
Silencing Specific Genes in Plants Using Virus-Induced Gene Silencing (VIGS) Vectors.Methods Mol Biol. 2022;2400:149-161. doi: 10.1007/978-1-0716-1835-6_15. Methods Mol Biol. 2022. PMID: 34905199
-
Foxtail mosaic virus: A tool for gene function analysis in maize and other monocots.Mol Plant Pathol. 2023 Jul;24(7):811-822. doi: 10.1111/mpp.13330. Epub 2023 Apr 10. Mol Plant Pathol. 2023. PMID: 37036421 Free PMC article. Review.
-
The potential of virus-induced gene silencing for speeding up functional characterization of plant genes.Genet Mol Res. 2004 Sep 30;3(3):323-41. Genet Mol Res. 2004. PMID: 15614725 Review.
Cited by
-
Development of a robust and efficient virus-induced gene silencing system for reverse genetics in recalcitrant Camellia drupifera capsules.Plant Methods. 2025 Jan 3;21(1):1. doi: 10.1186/s13007-024-01320-1. Plant Methods. 2025. PMID: 39754266 Free PMC article.
-
Virus-induced gene silencing for in planta validation of gene function in cucurbits.Plant Physiol. 2022 Nov 28;190(4):2366-2379. doi: 10.1093/plphys/kiac363. Plant Physiol. 2022. PMID: 35944218 Free PMC article.
-
Beyond salt tolerance: SOS1-13's pivotal role in regulating the immune response to Fusarium oxysporum in Solanum phureja.Front Plant Sci. 2025 Mar 6;16:1553348. doi: 10.3389/fpls.2025.1553348. eCollection 2025. Front Plant Sci. 2025. PMID: 40115954 Free PMC article.
-
Single-cell transcriptomic profiling of maize cell heterogeneity and systemic immune responses against Puccinia polysora Underw.Plant Biotechnol J. 2025 Feb;23(2):549-563. doi: 10.1111/pbi.14519. Epub 2024 Nov 29. Plant Biotechnol J. 2025. PMID: 39612313 Free PMC article.
-
Genome-Wide Analysis of C-Repeat Binding Factor Gene Family in Capsicum baccatum and Functional Exploration in Low-Temperature Response.Plants (Basel). 2024 Feb 17;13(4):549. doi: 10.3390/plants13040549. Plants (Basel). 2024. PMID: 38498531 Free PMC article.
References
-
- Becker A, Lange M (2010) VIGS—genomics goes functional. Trends Plant Sci 15:1–4 - PubMed
-
- Bernacki S, Karimi M, Hilson P, Robertson N (2010) Virus-induced gene silencing as a reverse genetics tool to study gene function. Methods Mol Biol 655:27–45 - PubMed
-
- Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP (2004) Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J 39:734–746 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

