ProNAB: database for binding affinities of protein-nucleic acid complexes and their mutants
- PMID: 34606614
- PMCID: PMC8728258
- DOI: 10.1093/nar/gkab848
ProNAB: database for binding affinities of protein-nucleic acid complexes and their mutants
Abstract
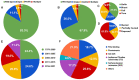
Protein-nucleic acid interactions are involved in various biological processes such as gene expression, replication, transcription, translation and packaging. The binding affinities of protein-DNA and protein-RNA complexes are important for elucidating the mechanism of protein-nucleic acid recognition. Although experimental data on binding affinity are reported abundantly in the literature, no well-curated database is currently available for protein-nucleic acid binding affinity. We have developed a database, ProNAB, which contains more than 20 000 experimental data for the binding affinities of protein-DNA and protein-RNA complexes. Each entry provides comprehensive information on sequence and structural features of a protein, nucleic acid and its complex, experimental conditions, thermodynamic parameters such as dissociation constant (Kd), binding free energy (ΔG) and change in binding free energy upon mutation (ΔΔG), and literature information. ProNAB is cross-linked with GenBank, UniProt, PDB, ProThermDB, PROSITE, DisProt and Pubmed. It provides a user-friendly web interface with options for search, display, sorting, visualization, download and upload the data. ProNAB is freely available at https://web.iitm.ac.in/bioinfo2/pronab/ and it has potential applications such as understanding the factors influencing the affinity, development of prediction tools, binding affinity change upon mutation and design complexes with the desired affinity.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
MPAD: A Database for Binding Affinity of Membrane Protein-protein Complexes and their Mutants.J Mol Biol. 2023 Jul 15;435(14):167870. doi: 10.1016/j.jmb.2022.167870. Epub 2022 Oct 26. J Mol Biol. 2023. PMID: 36309134
-
Thermodynamic database for protein-nucleic acid interactions (ProNIT).Bioinformatics. 2001 Nov;17(11):1027-34. doi: 10.1093/bioinformatics/17.11.1027. Bioinformatics. 2001. PMID: 11724731
-
ProCaff: protein-carbohydrate complex binding affinity database.Bioinformatics. 2020 Jun 1;36(11):3615-3617. doi: 10.1093/bioinformatics/btaa141. Bioinformatics. 2020. PMID: 32119071
-
Protein-nucleic acid complexes: Docking and binding affinity.Curr Opin Struct Biol. 2025 Feb;90:102955. doi: 10.1016/j.sbi.2024.102955. Epub 2024 Nov 30. Curr Opin Struct Biol. 2025. PMID: 39616716 Review.
-
Thermodynamic databases for proteins and protein-nucleic acid interactions.Biopolymers. 2001-2002;61(2):121-6. doi: 10.1002/1097-0282(2002)61:2<121::AID-BIP10077>3.0.CO;2-1. Biopolymers. 2001. PMID: 11987161 Review.
Cited by
-
Stimulus-Responsive Nanodelivery and Release Systems for Cancer Gene Therapy: Efficacy Improvement Strategies.Int J Nanomedicine. 2024 Jul 12;19:7099-7121. doi: 10.2147/IJN.S470637. eCollection 2024. Int J Nanomedicine. 2024. PMID: 39045344 Free PMC article. Review.
-
Bioinformatics Approaches for Understanding the Binding Affinity of Protein-Nucleic Acid Complexes.Methods Mol Biol. 2025;2867:315-330. doi: 10.1007/978-1-0716-4196-5_18. Methods Mol Biol. 2025. PMID: 39576589
-
Overview and limitations of database in global traditional medicines: A narrative review.Acta Pharmacol Sin. 2025 Feb;46(2):235-263. doi: 10.1038/s41401-024-01353-1. Epub 2024 Aug 2. Acta Pharmacol Sin. 2025. PMID: 39095509 Review.
-
Large-Scale Multi-omic Biosequence Transformers for Modeling Protein-Nucleic Acid Interactions.ArXiv [Preprint]. 2025 Jun 18:arXiv:2408.16245v5. ArXiv. 2025. PMID: 40236839 Free PMC article. Preprint.
-
DNAPred_Prot: Identification of DNA-Binding Proteins Using Composition- and Position-Based Features.Appl Bionics Biomech. 2022 Apr 13;2022:5483115. doi: 10.1155/2022/5483115. eCollection 2022. Appl Bionics Biomech. 2022. PMID: 35465187 Free PMC article.
References
-
- Crocker J., Noon E.P., Stern D.L.. The soft touch: low-affinity transcription factor binding sites in development and evolution. Curr. Top. Dev. Biol. 2016; 117:455–469. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

