Taxon-specific or universal? Using target capture to study the evolutionary history of rapid radiations
- PMID: 34606683
- PMCID: PMC9292372
- DOI: 10.1111/1755-0998.13523
Taxon-specific or universal? Using target capture to study the evolutionary history of rapid radiations
Abstract
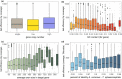
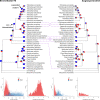
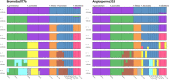
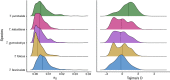
Target capture has emerged as an important tool for phylogenetics and population genetics in nonmodel taxa. Whereas developing taxon-specific capture probes requires sustained efforts, available universal kits may have a lower power to reconstruct relationships at shallow phylogenetic scales and within rapidly radiating clades. We present here a newly developed target capture set for Bromeliaceae, a large and ecologically diverse plant family with highly variable diversification rates. The set targets 1776 coding regions, including genes putatively involved in key innovations, with the aim to empower testing of a wide range of evolutionary hypotheses. We compare the relative power of this taxon-specific set, Bromeliad1776, to the universal Angiosperms353 kit. The taxon-specific set results in higher enrichment success across the entire family; however, the overall performance of both kits to reconstruct phylogenetic trees is relatively comparable, highlighting the vast potential of universal kits for resolving evolutionary relationships. For more detailed phylogenetic or population genetic analyses, for example the exploration of gene tree concordance, nucleotide diversity or population structure, the taxon-specific capture set presents clear benefits. We discuss the potential lessons that this comparative study provides for future phylogenetic and population genetic investigations, in particular for the study of evolutionary radiations.
La captura selectiva de secuencias de ADN ha surgido como una herramienta importante para la filogenética y la genética de poblaciones en taxones no-modelo. Mientras que el desarrollo de sondas de captura específicas para cada taxón requiere un esfuerzo sostenido, las colecciones de sondas universales disponibles pueden tener una potencia disminuida para la reconstrucción de relaciones filogenéticas poco profundas o de radiaciones rápidas. Presentamos aquí un conjunto de sondas para la captura selectiva desarrollado recientemente para Bromeliaceae, una familia de plantas extensa, ecológicamente diversa y con tasas de diversificación muy variables. El conjunto de sondas se centra en 1776 regiones de codificación, incluyendo genes supuestamente implicados en rasgos de innovación clave, con el objetivo de potenciar la comprobación de una amplia gama de hipótesis evolutivas. Comparamos la potencia relativa de este conjunto de sondas diseñado para un taxón específico, Bromeliad1776, con la colección universal Angiosperms353. El conjunto específico da lugar a un mayor éxito de captura en toda la familia. Sin embargo, el rendimiento global de ambos kits para reconstruir árboles filogenéticos es relativamente comparable, lo que pone de manifiesto el gran potencial de los kits universales para resolver las relaciones evolutivas. Para análisis filogenéticos o de genética de poblaciones más detallados, como por ejemplo la exploración de la congruencia de los árboles de genes, la diversidad de nucleótidos o la estructura de la población, el conjunto de captura específico para Bromeliaceae presenta claras ventajas. Discutimos las lecciones potenciales que este estudio comparativo proporciona para futuras investigaciones filogenéticas y de genética de poblaciones, en particular para el estudio de las radiaciones evolutivas.
Keywords: Tillandsia; Bromeliaceae; phylogenomics; plant radiation; population structure; target capture.
© 2021 The Authors. Molecular Ecology Resources published by John Wiley & Sons Ltd.
Figures

Similar articles
-
Tackling Rapid Radiations With Targeted Sequencing.Front Plant Sci. 2020 Jan 9;10:1655. doi: 10.3389/fpls.2019.01655. eCollection 2019. Front Plant Sci. 2020. PMID: 31998342 Free PMC article.
-
Utility of targeted sequence capture for phylogenomics in rapid, recent angiosperm radiations: Neotropical Burmeistera bellflowers as a case study.Mol Phylogenet Evol. 2020 Nov;152:106769. doi: 10.1016/j.ympev.2020.106769. Epub 2020 Feb 17. Mol Phylogenet Evol. 2020. PMID: 32081762
-
Target-capture probes for phylogenomics of the Caenogastropoda.Mol Ecol Resour. 2023 Aug;23(6):1372-1388. doi: 10.1111/1755-0998.13793. Epub 2023 Apr 9. Mol Ecol Resour. 2023. PMID: 36997300
-
The genetics of evolutionary radiations.Biol Rev Camb Philos Soc. 2020 Aug;95(4):1055-1072. doi: 10.1111/brv.12598. Epub 2020 Mar 31. Biol Rev Camb Philos Soc. 2020. PMID: 32233014 Review.
-
Confluence, synnovation, and depauperons in plant diversification.New Phytol. 2015 Jul;207(2):260-274. doi: 10.1111/nph.13367. Epub 2015 Mar 16. New Phytol. 2015. PMID: 25778694 Review.
Cited by
-
A roadmap of phylogenomic methods for studying polyploid plant genera.Appl Plant Sci. 2024 Apr 22;12(4):e11580. doi: 10.1002/aps3.11580. eCollection 2024 Jul-Aug. Appl Plant Sci. 2024. PMID: 39184196 Free PMC article.
-
Genetic Drift Shapes the Evolution of a Highly Dynamic Metapopulation.Mol Biol Evol. 2022 Dec 5;39(12):msac264. doi: 10.1093/molbev/msac264. Mol Biol Evol. 2022. PMID: 36472514 Free PMC article.
-
A target enrichment probe set for resolving phylogenetic relationships in the coffee family, Rubiaceae.Appl Plant Sci. 2023 Nov 17;11(6):e11554. doi: 10.1002/aps3.11554. eCollection 2023 Nov-Dec. Appl Plant Sci. 2023. PMID: 38106541 Free PMC article.
-
Evaluating the utility of deep genome skimming for phylogenomic analyses: A case study in the species-rich genus Rhododendron.Plant Divers. 2025 May 2;47(4):593-603. doi: 10.1016/j.pld.2025.04.006. eCollection 2025 Jul. Plant Divers. 2025. PMID: 40734829 Free PMC article.
-
Developing Asparagaceae1726: An Asparagaceae-specific probe set targeting 1726 loci for Hyb-Seq and phylogenomics in the family.Appl Plant Sci. 2024 Jun 18;12(5):e11597. doi: 10.1002/aps3.11597. eCollection 2024 Sep-Oct. Appl Plant Sci. 2024. PMID: 39360194 Free PMC article.
References
-
- Aguirre‐Santoro, J. , Salinas, N. R. , & Michelangeli, F. A. (2020). The influence of floral variation and geographic disjunction on the evolutionary dynamics of Ronnbergia and Wittmackia (Bromeliaceae: Bromelioideae). Botanical Journal of the Linnean Society, 192(4), 609–624. 10.1093/botlinnean/boz087 - DOI
-
- Andrews, S. (2010). FastQC: A quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute.
-
- Bagley, J. C. , Uribe‐Convers, S. , Carlsen, M. M. , & Muchhala, N. (2020). Utility of targeted sequence capture for phylogenomics in rapid, recent angiosperm radiations: Neotropical Burmeistera bellflowers as a case study. Molecular Phylogenetics and Evolution, 152, 106769. 10.1016/j.ympev.2020.106769 - DOI - PubMed
-
- Barfuss, M. H. , Till, W. , Leme, E. M. , Pinzón, J. P. , Manzanares, J. M. , Halbritter, H. , Samuel, R. , & Brown, G. K. (2016). Taxonomic revision of Bromeliaceae subfam. Tillandsioideae based on a multi‐locus DNA sequence phylogeny and morphology. Phytotaxa, 279(1), 1–97. 10.11646/phytotaxa.279.1.1 - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

