Identification, visualization, statistical analysis and mathematical modeling of high-feedback loops in gene regulatory networks
- PMID: 34607562
- PMCID: PMC8489061
- DOI: 10.1186/s12859-021-04405-z
Identification, visualization, statistical analysis and mathematical modeling of high-feedback loops in gene regulatory networks
Abstract
Background: Feedback loops in gene regulatory networks play pivotal roles in governing functional dynamics of cells. Systems approaches demonstrated characteristic dynamical features, including multistability and oscillation, of positive and negative feedback loops. Recent experiments and theories have implicated highly interconnected feedback loops (high-feedback loops) in additional nonintuitive functions, such as controlling cell differentiation rate and multistep cell lineage progression. However, it remains challenging to identify and visualize high-feedback loops in complex gene regulatory networks due to the myriad of ways in which the loops can be combined. Furthermore, it is unclear whether the high-feedback loop structures with these potential functions are widespread in biological systems. Finally, it remains challenging to understand diverse dynamical features, such as high-order multistability and oscillation, generated by individual networks containing high-feedback loops. To address these problems, we developed HiLoop, a toolkit that enables discovery, visualization, and analysis of several types of high-feedback loops in large biological networks.
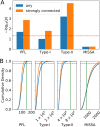
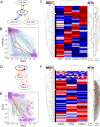
Results: HiLoop not only extracts high-feedback structures and visualize them in intuitive ways, but also quantifies the enrichment of overrepresented structures. Through random parameterization of mathematical models derived from target networks, HiLoop presents characteristic features of the underlying systems, including complex multistability and oscillations, in a unifying framework. Using HiLoop, we were able to analyze realistic gene regulatory networks containing dozens to hundreds of genes, and to identify many small high-feedback systems. We found more than a 100 human transcription factors involved in high-feedback loops that were not studied previously. In addition, HiLoop enabled the discovery of an enrichment of high feedback in pathways related to epithelial-mesenchymal transition.
Conclusions: HiLoop makes the study of complex networks accessible without significant computational demands. It can serve as a hypothesis generator through identification and modeling of high-feedback subnetworks, or as a quantification method for motif enrichment analysis. As an example of discovery, we found that multistep cell lineage progression may be driven by either specific instances of high-feedback loops with sparse appearances, or generally enriched topologies in gene regulatory networks. We expect HiLoop's usefulness to increase as experimental data of regulatory networks accumulate. Code is freely available for use or extension at https://github.com/BenNordick/HiLoop .
Keywords: Epithelial-mesenchymal transition; Feedback loop; Gene regulatory network; Multistability; Oscillation; T cell differentiation.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Multistability, oscillations and bifurcations in feedback loops.Math Biosci Eng. 2010 Jan;7(1):83-97. doi: 10.3934/mbe.2010.7.83. Math Biosci Eng. 2010. PMID: 20104950
-
A synthetic gene circuit for measuring autoregulatory feedback control.Integr Biol (Camb). 2016 Apr 18;8(4):546-55. doi: 10.1039/c5ib00230c. Epub 2016 Jan 5. Integr Biol (Camb). 2016. PMID: 26728081
-
Coupling between feedback loops in autoregulatory networks affects bistability range, open-loop gain and switching times.Phys Biol. 2012 Oct;9(5):055003. doi: 10.1088/1478-3975/9/5/055003. Epub 2012 Sep 25. Phys Biol. 2012. PMID: 23011599 Free PMC article.
-
Design principles of regulatory networks underlying epithelial mesenchymal plasticity in cancer cells.Curr Opin Cell Biol. 2025 Feb;92:102445. doi: 10.1016/j.ceb.2024.102445. Epub 2024 Nov 27. Curr Opin Cell Biol. 2025. PMID: 39608060 Review.
-
Oscillation patterns in negative feedback loops.Proc Natl Acad Sci U S A. 2007 Apr 17;104(16):6533-7. doi: 10.1073/pnas.0610759104. Epub 2007 Apr 5. Proc Natl Acad Sci U S A. 2007. PMID: 17412833 Free PMC article. Review.
Cited by
-
Genome-wide inference reveals that feedback regulations constrain promoter-dependent transcriptional burst kinetics.Nucleic Acids Res. 2023 Jan 11;51(1):68-83. doi: 10.1093/nar/gkac1204. Nucleic Acids Res. 2023. PMID: 36583343 Free PMC article.
-
Operating principles of interconnected feedback loops driving cell fate transitions.NPJ Syst Biol Appl. 2025 Jan 2;11(1):2. doi: 10.1038/s41540-024-00483-w. NPJ Syst Biol Appl. 2025. PMID: 39743534 Free PMC article.
-
Cooperative RNA degradation stabilizes intermediate epithelial-mesenchymal states and supports a phenotypic continuum.iScience. 2022 Sep 27;25(10):105224. doi: 10.1016/j.isci.2022.105224. eCollection 2022 Oct 21. iScience. 2022. PMID: 36248730 Free PMC article.
-
A quantitative evaluation of topological motifs and their coupling in gene circuit state distributions.iScience. 2023 Jan 23;26(2):106029. doi: 10.1016/j.isci.2023.106029. eCollection 2023 Feb 17. iScience. 2023. PMID: 36824273 Free PMC article.
-
Network topology metrics explaining enrichment of hybrid epithelial/mesenchymal phenotypes in metastasis.PLoS Comput Biol. 2022 Nov 8;18(11):e1010687. doi: 10.1371/journal.pcbi.1010687. eCollection 2022 Nov. PLoS Comput Biol. 2022. PMID: 36346808 Free PMC article.
References
-
- Yao G, Lee TJ, Mori S, Nevins JR, You L. A bistable Rb–E2F switch underlies the restriction point. Nat Cell Biol. 2008;10(4):476. - PubMed
-
- Zhang J, Tian XJ, Zhang H, Teng Y, Li R, Bai F, Elankumaran S, Xing J. TGF-β -induced epithelial-to-mesenchymal transition proceeds through stepwise activation of multiple feedback loops. Sci Signal. 2014;7(345):ra91. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

