LPI-deepGBDT: a multiple-layer deep framework based on gradient boosting decision trees for lncRNA-protein interaction identification
- PMID: 34607567
- PMCID: PMC8489074
- DOI: 10.1186/s12859-021-04399-8
LPI-deepGBDT: a multiple-layer deep framework based on gradient boosting decision trees for lncRNA-protein interaction identification
Abstract
Background: Long noncoding RNAs (lncRNAs) play important roles in various biological and pathological processes. Discovery of lncRNA-protein interactions (LPIs) contributes to understand the biological functions and mechanisms of lncRNAs. Although wet experiments find a few interactions between lncRNAs and proteins, experimental techniques are costly and time-consuming. Therefore, computational methods are increasingly exploited to uncover the possible associations. However, existing computational methods have several limitations. First, majority of them were measured based on one simple dataset, which may result in the prediction bias. Second, few of them are applied to identify relevant data for new lncRNAs (or proteins). Finally, they failed to utilize diverse biological information of lncRNAs and proteins.
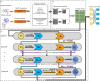
Results: Under the feed-forward deep architecture based on gradient boosting decision trees (LPI-deepGBDT), this work focuses on classify unobserved LPIs. First, three human LPI datasets and two plant LPI datasets are arranged. Second, the biological features of lncRNAs and proteins are extracted by Pyfeat and BioProt, respectively. Thirdly, the features are dimensionally reduced and concatenated as a vector to represent an lncRNA-protein pair. Finally, a deep architecture composed of forward mappings and inverse mappings is developed to predict underlying linkages between lncRNAs and proteins. LPI-deepGBDT is compared with five classical LPI prediction models (LPI-BLS, LPI-CatBoost, PLIPCOM, LPI-SKF, and LPI-HNM) under three cross validations on lncRNAs, proteins, lncRNA-protein pairs, respectively. It obtains the best average AUC and AUPR values under the majority of situations, significantly outperforming other five LPI identification methods. That is, AUCs computed by LPI-deepGBDT are 0.8321, 0.6815, and 0.9073, respectively and AUPRs are 0.8095, 0.6771, and 0.8849, respectively. The results demonstrate the powerful classification ability of LPI-deepGBDT. Case study analyses show that there may be interactions between GAS5 and Q15717, RAB30-AS1 and O00425, and LINC-01572 and P35637.
Conclusions: Integrating ensemble learning and hierarchical distributed representations and building a multiple-layered deep architecture, this work improves LPI prediction performance as well as effectively probes interaction data for new lncRNAs/proteins.
Keywords: Gradient boosting decision tree; Multiple-layer deep architecture; lncRNA–protein interaction.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
LPI-HyADBS: a hybrid framework for lncRNA-protein interaction prediction integrating feature selection and classification.BMC Bioinformatics. 2021 Nov 26;22(1):568. doi: 10.1186/s12859-021-04485-x. BMC Bioinformatics. 2021. PMID: 34836494 Free PMC article.
-
LPI-EnEDT: an ensemble framework with extra tree and decision tree classifiers for imbalanced lncRNA-protein interaction data classification.BioData Min. 2021 Dec 3;14(1):50. doi: 10.1186/s13040-021-00277-4. BioData Min. 2021. PMID: 34861891 Free PMC article.
-
EnANNDeep: An Ensemble-based lncRNA-protein Interaction Prediction Framework with Adaptive k-Nearest Neighbor Classifier and Deep Models.Interdiscip Sci. 2022 Mar;14(1):209-232. doi: 10.1007/s12539-021-00483-y. Epub 2022 Jan 10. Interdiscip Sci. 2022. PMID: 35006529
-
Probing lncRNA-Protein Interactions: Data Repositories, Models, and Algorithms.Front Genet. 2020 Jan 31;10:1346. doi: 10.3389/fgene.2019.01346. eCollection 2019. Front Genet. 2020. PMID: 32082358 Free PMC article. Review.
-
LMI-DForest: A deep forest model towards the prediction of lncRNA-miRNA interactions.Comput Biol Chem. 2020 Dec;89:107406. doi: 10.1016/j.compbiolchem.2020.107406. Epub 2020 Oct 20. Comput Biol Chem. 2020. PMID: 33120126 Review.
Cited by
-
LncRNA-Top: Controlled deep learning approaches for lncRNA gene regulatory relationship annotations across different platforms.iScience. 2023 Oct 12;26(11):108197. doi: 10.1016/j.isci.2023.108197. eCollection 2023 Nov 17. iScience. 2023. PMID: 37965148 Free PMC article.
-
Long-distance dependency combined multi-hop graph neural networks for protein-protein interactions prediction.BMC Bioinformatics. 2022 Dec 5;23(1):521. doi: 10.1186/s12859-022-05062-6. BMC Bioinformatics. 2022. PMID: 36471248 Free PMC article.
-
Predicting lncRNA-protein interactions through deep learning framework employing multiple features and random forest algorithm.BMC Bioinformatics. 2024 Mar 12;25(1):108. doi: 10.1186/s12859-024-05727-4. BMC Bioinformatics. 2024. PMID: 38475723 Free PMC article.
-
Combining a machine-learning derived 4-lncRNA signature with AFP and TNM stages in predicting early recurrence of hepatocellular carcinoma.BMC Genomics. 2023 Feb 27;24(1):89. doi: 10.1186/s12864-023-09194-8. BMC Genomics. 2023. PMID: 36849926 Free PMC article.
-
Predicting circRNA-drug sensitivity associations via graph attention auto-encoder.BMC Bioinformatics. 2022 May 4;23(1):160. doi: 10.1186/s12859-022-04694-y. BMC Bioinformatics. 2022. PMID: 35508967 Free PMC article.
References
-
- Liu Z-P. Predicting lncrna-protein interactions by machine learning methods: a review. Curr. Bioinform. 2020;15(8):831–840.
-
- Chen X, Sun Y-Z, Guan N-N, Qu J, Huang Z-A, Zhu Z-X, Li J-Q. Computational models for lncrna function prediction and functional similarity calculation. Brief. Funct. Genom. 2019;18(1):58–82. - PubMed
-
- Wang, W., Dai, Q., Li, F., Xiong, Y., Wei, D.-Q.: Mlcdforest: multi-label classification with deep forest in disease prediction for long non-coding rnas. Brief. Bioinform. (2020) - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

