Hamster organotypic modeling of SARS-CoV-2 lung and brainstem infection
- PMID: 34608167
- PMCID: PMC8490365
- DOI: 10.1038/s41467-021-26096-z
Hamster organotypic modeling of SARS-CoV-2 lung and brainstem infection
Abstract
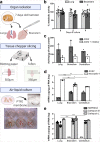
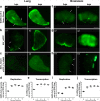
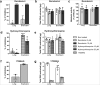
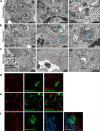
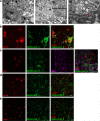
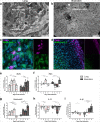
SARS-CoV-2 has caused a global pandemic of COVID-19 since its emergence in December 2019. The infection causes a severe acute respiratory syndrome and may also spread to central nervous system leading to neurological sequelae. We have developed and characterized two new organotypic cultures from hamster brainstem and lung tissues that offer a unique opportunity to study the early steps of viral infection and screening antivirals. These models are not dedicated to investigate how the virus reaches the brain. However, they allow validating the early tropism of the virus in the lungs and demonstrating that SARS-CoV-2 could infect the brainstem and the cerebellum, mainly by targeting granular neurons. Viral infection induces specific interferon and innate immune responses with patterns specific to each organ, along with cell death by apoptosis, necroptosis, and pyroptosis. Overall, our data illustrate the potential of rapid modeling of complex tissue-level interactions during infection by a newly emerged virus.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Worldometer. Coronavirus Cases. Worldometer (2021). Available at: https://www.worldometers.info/coronavirus/. (Accessed: 20th April 2021).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

