Natural diversity provides a broad spectrum of cyanobacteriochrome-based diguanylate cyclases
- PMID: 34608946
- PMCID: PMC8491021
- DOI: 10.1093/plphys/kiab240
Natural diversity provides a broad spectrum of cyanobacteriochrome-based diguanylate cyclases
Abstract
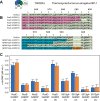
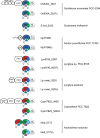
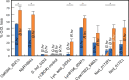
Cyanobacteriochromes (CBCRs) are spectrally diverse photosensors from cyanobacteria distantly related to phytochromes that exploit photoisomerization of linear tetrapyrrole (bilin) chromophores to regulate associated signaling output domains. Unlike phytochromes, a single CBCR domain is sufficient for photoperception. CBCR domains that regulate the production or degradation of cyclic nucleotide second messengers are becoming increasingly well characterized. Cyclic di-guanosine monophosphate (c-di-GMP) is a widespread small-molecule regulator of bacterial motility, developmental transitions, and biofilm formation whose biosynthesis is regulated by CBCRs coupled to GGDEF (diguanylate cyclase) output domains. In this study, we compare the properties of diverse CBCR-GGDEF proteins with those of synthetic CBCR-GGDEF chimeras. Our investigation shows that natural diversity generates promising candidates for robust, broad spectrum optogenetic applications in live cells. Since light quality is constantly changing during plant development as upper leaves begin to shade lower leaves-affecting elongation growth, initiation of flowering, and responses to pathogens, these studies presage application of CBCR-GGDEF sensors to regulate orthogonal, c-di-GMP-regulated circuits in agronomically important plants for robust mitigation of such deleterious responses under natural growing conditions in the field.
© American Society of Plant Biologists 2021. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures



References
-
- Anders K, Essen LO (2015) The family of phytochrome-like photoreceptors: diverse, complex and multi-colored, but very useful. Curr Opin Struct Biol 35: 7–16 - PubMed
-
- Beattie GA, Hatfield BM, Dong HL, McGrane RS (2018) Seeing the light: The roles of red- and blue-light sensing in plant microbes. Annu Rev Phytopathol 56: 41–66 - PubMed
-
- Blain-Hartung MD, Rockwell NC, Lagarias JC (2017) Light-regulated synthesis of cyclic-di-GMP by a bidomain construct of the cyanobacteriochrome Tlr0924 (SesA) without stable dimerization. Biochemistry 56: 6145–6154 - PubMed
-
- Burgie ES, Walker JM, Phillips GN Jr, Vierstra RD (2013) A photo-labile thioether linkage to phycoviolobilin provides the foundation for the blue/green photocycles in DXCF-cyanobacteriochromes. Structure 21: 88–97 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

