A nitrogen stress-inducible small RNA regulates CO2 fixation in Nostoc
- PMID: 34608966
- PMCID: PMC8491059
- DOI: 10.1093/plphys/kiab309
A nitrogen stress-inducible small RNA regulates CO2 fixation in Nostoc
Abstract
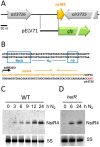
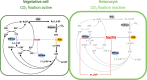
In the absence of fixed nitrogen, some filamentous cyanobacteria differentiate heterocysts, specialized cells devoted to fixing atmospheric nitrogen (N2). This differentiation process is controlled by the global nitrogen regulator NtcA and involves extensive metabolic reprogramming, including shutdown of photosynthetic CO2 fixation in heterocysts, to provide a microaerobic environment suitable for N2 fixation. Small regulatory RNAs (sRNAs) are major post-transcriptional regulators of gene expression in bacteria. In cyanobacteria, responding to nitrogen deficiency involves transcribing several nitrogen-regulated sRNAs. Here, we describe the participation of nitrogen stress-inducible RNA 4 (NsiR4) in post-transcriptionally regulating the expression of two genes involved in CO2 fixation via the Calvin cycle: glpX, which encodes bifunctional sedoheptulose-1,7-bisphosphatase/fructose-1,6-bisphosphatase (SBPase), and pgk, which encodes phosphoglycerate kinase (PGK). Using a heterologous reporter assay in Escherichia coli, we show that NsiR4 interacts with the 5'-untranslated region (5'-UTR) of glpX and pgk mRNAs. Overexpressing NsiR4 in Nostoc sp. PCC 7120 resulted in a reduced amount of SBPase protein and reduced PGK activity, as well as reduced levels of both glpX and pgk mRNAs, further supporting that NsiR4 negatively regulates these two enzymes. In addition, using a gfp fusion to the nsiR4 promoter, we show stronger expression of NsiR4 in heterocysts than in vegetative cells, which could contribute to the heterocyst-specific shutdown of Calvin cycle flux. Post-transcriptional regulation of two Calvin cycle enzymes by NsiR4, a nitrogen-regulated sRNA, represents an additional link between nitrogen control and CO2 assimilation.
© The Author(s) 2021. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Figures



Similar articles
-
A Heterocyst-Specific Antisense RNA Contributes to Metabolic Reprogramming in Nostoc sp. PCC 7120.Plant Cell Physiol. 2019 Aug 1;60(8):1646-1655. doi: 10.1093/pcp/pcz087. Plant Cell Physiol. 2019. PMID: 31093664
-
NsiR3, a nitrogen stress-inducible small RNA, regulates proline oxidase expression in the cyanobacterium Nostoc sp. PCC 7120.FEBS J. 2021 Mar;288(5):1614-1629. doi: 10.1111/febs.15516. Epub 2020 Sep 2. FEBS J. 2021. PMID: 32799414
-
The Nitrogen Stress-Repressed sRNA NsrR1 Regulates Expression of all1871, a Gene Required for Diazotrophic Growth in Nostoc sp. PCC 7120.Life (Basel). 2020 Apr 29;10(5):54. doi: 10.3390/life10050054. Life (Basel). 2020. PMID: 32365616 Free PMC article.
-
Genetic responses to carbon and nitrogen availability in Anabaena.Environ Microbiol. 2019 Jan;21(1):1-17. doi: 10.1111/1462-2920.14370. Epub 2018 Oct 16. Environ Microbiol. 2019. PMID: 30066380 Review.
-
Transcriptional regulation of development in heterocyst-forming cyanobacteria.Biochim Biophys Acta Gene Regul Mech. 2019 Jul;1862(7):673-684. doi: 10.1016/j.bbagrm.2018.04.006. Epub 2018 Apr 30. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 29719238 Review.
Cited by
-
Gene network centrality analysis identifies key regulators coordinating day-night metabolic transitions in Synechococcus elongatus PCC 7942 despite limited accuracy in predicting direct regulator-gene interactions.Front Microbiol. 2025 Mar 26;16:1569559. doi: 10.3389/fmicb.2025.1569559. eCollection 2025. Front Microbiol. 2025. PMID: 40207147 Free PMC article.
-
The Heterocyst-Specific Small RNA NsiR1 Regulates the Commitment to Differentiation in Nostoc.Microbiol Spectr. 2022 Apr 27;10(2):e0227421. doi: 10.1128/spectrum.02274-21. Epub 2022 Mar 1. Microbiol Spectr. 2022. PMID: 35230129 Free PMC article.
-
The sRNA NsiR4 fine-tunes arginine synthesis in the cyanobacterium Synechocystis sp. PCC 6803 by post-transcriptional regulation of PirA.RNA Biol. 2022 Jan;19(1):811-818. doi: 10.1080/15476286.2022.2082147. RNA Biol. 2022. PMID: 35678613 Free PMC article.
-
Antisense RNA regulates glutamine synthetase in a heterocyst-forming cyanobacterium.Plant Physiol. 2024 Jul 31;195(4):2911-2920. doi: 10.1093/plphys/kiae263. Plant Physiol. 2024. PMID: 38708585 Free PMC article.
-
Molecular Mechanisms of Nostoc flagelliforme Environmental Adaptation: A Comprehensive Review.Plants (Basel). 2025 May 22;14(11):1582. doi: 10.3390/plants14111582. Plants (Basel). 2025. PMID: 40508256 Free PMC article. Review.
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 - PubMed
-
- Álvarez-Escribano I, Brenes-Álvarez M, Olmedo-Verd E, Georg J, Hess WR, Vioque A, Muro-Pastor AM (2021) NsiR3, a nitrogen stress-inducible small RNA, regulates proline oxidase expression in the cyanobacterium Nostoc sp. PCC 7120. FEBS J 288: 1614–1629 - PubMed
-
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 - PubMed
-
- Brenes-Álvarez M, Minguet M, Vioque A, Muro-Pastor AM (2020a. ) NsiR1, a small RNA with multiple copies, modulates heterocyst differentiation in the cyanobacterium Nostoc sp. PCC 7120. Environ Microbiol 22: 3325–3338 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

