Family-wide evaluation of RAPID ALKALINIZATION FACTOR peptides
- PMID: 34608971
- PMCID: PMC8491022
- DOI: 10.1093/plphys/kiab308
Family-wide evaluation of RAPID ALKALINIZATION FACTOR peptides
Abstract
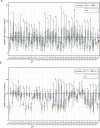
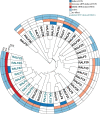
Plant peptide hormones are important players that control various aspects of the lives of plants. RAPID ALKALINIZATION FACTOR (RALF) peptides have recently emerged as important players in multiple physiological processes. Numerous studies have increased our understanding of the evolutionary processes that shaped the RALF family of peptides. Nevertheless, to date, there is no comprehensive, family-wide functional study on RALF peptides. Here, we analyzed the phylogeny of the proposed multigenic RALF peptide family in the model plant Arabidopsis (Arabidopsis thaliana), ecotype Col-0, and tested a variety of physiological responses triggered by RALFs. Our phylogenetic analysis reveals that two of the previously proposed RALF peptides are not genuine RALF peptides, which leads us to propose a revision to the consensus AtRALF peptide family annotation. We show that the majority of AtRALF peptides, when applied exogenously as synthetic peptides, induce seedling or root growth inhibition and modulate reactive oxygen species (ROS) production in Arabidopsis. Moreover, our findings suggest that alkalinization and growth inhibition are, generally, coupled characteristics of RALF peptides. Additionally, we show that for the majority of the peptides, these responses are genetically dependent on FERONIA, suggesting a pivotal role for this receptor kinase in the perception of multiple RALF peptides.
© The Author(s) 2021. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

