Deep Learning Coordinate-Free Quantum Chemistry
- PMID: 34609871
- PMCID: PMC10348818
- DOI: 10.1021/acs.jpca.1c04462
Deep Learning Coordinate-Free Quantum Chemistry
Abstract
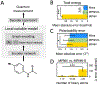
Computing quantum chemical properties of small molecules and polymers can provide insights valuable into physicists, chemists, and biologists when designing new materials, catalysts, biological probes, and drugs. Deep learning can compute quantum chemical properties accurately in a fraction of time required by commonly used methods such as density functional theory. Most current approaches to deep learning in quantum chemistry begin with geometric information from experimentally derived molecular structures or pre-calculated atom coordinates. These approaches have many useful applications, but they can be costly in time and computational resources. In this study, we demonstrate that accurate quantum chemical computations can be performed without geometric information by operating in the coordinate-free domain using deep learning on graph encodings. Coordinate-free methods rely only on molecular graphs, the connectivity of atoms and bonds, without atom coordinates or bond distances. We also find that the choice of graph-encoding architecture substantially affects the performance of these methods. The structures of these graph-encoding architectures provide an opportunity to probe an important, outstanding question in quantum mechanics: what types of quantum chemical properties can be represented by local variable models? We find that Wave, a local variable model, accurately calculates the quantum chemical properties, while graph convolutional architectures require global variables. Furthermore, local variable Wave models outperform global variable graph convolution models on complex molecules with large, correlated systems.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Segler MHS; Preuss M; Waller MP Planning chemical syntheses with deep neural networks and symbolic AI. Nature 2018, 555, 604–610. - PubMed
-
- Titano JJ; Badgeley M; Schefflein J; Pain M; Su A; Cai M; Swinburne N; Zech J; Kim J; Bederson J; et al. Automated deep-neural-network surveillance of cranial images for acute neurologic events. Nat. Med 2018, 24, 1337–1341. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

