Controlled human malaria infection with a clone of Plasmodium vivax with high-quality genome assembly
- PMID: 34609964
- PMCID: PMC8675201
- DOI: 10.1172/jci.insight.152465
Controlled human malaria infection with a clone of Plasmodium vivax with high-quality genome assembly
Abstract
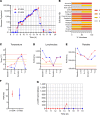
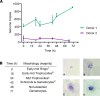
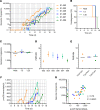
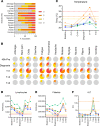
Controlled human malaria infection (CHMI) provides a highly informative means to investigate host-pathogen interactions and enable in vivo proof-of-concept efficacy testing of new drugs and vaccines. However, unlike Plasmodium falciparum, well-characterized P. vivax parasites that are safe and suitable for use in modern CHMI models are limited. Here, 2 healthy malaria-naive United Kingdom adults with universal donor blood group were safely infected with a clone of P. vivax from Thailand by mosquito-bite CHMI. Parasitemia developed in both volunteers, and prior to treatment, each volunteer donated blood to produce a cryopreserved stabilate of infected RBCs. Following stringent safety screening, the parasite stabilate from one of these donors (PvW1) was thawed and used to inoculate 6 healthy malaria-naive United Kingdom adults by blood-stage CHMI, at 3 different dilutions. Parasitemia developed in all volunteers, who were then successfully drug treated. PvW1 parasite DNA was isolated and sequenced to produce a high-quality genome assembly by using a hybrid assembly method. We analyzed leading vaccine candidate antigens and multigene families, including the vivax interspersed repeat (VIR) genes, of which we identified 1145 in the PvW1 genome. Our genomic analysis will guide future assessment of candidate vaccines and drugs, as well as experimental medicine studies.
Trial registration: ClinicalTrials.gov NCT03797989 NCT03377296.
Keywords: Infectious disease; Malaria.
Figures

References
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

