RBMS1 regulates lung cancer ferroptosis through translational control of SLC7A11
- PMID: 34609966
- PMCID: PMC8592553
- DOI: 10.1172/JCI152067
RBMS1 regulates lung cancer ferroptosis through translational control of SLC7A11
Abstract
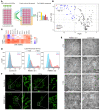
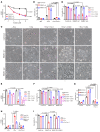
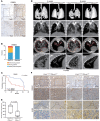
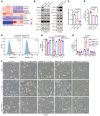
Ferroptosis, an iron-dependent nonapoptotic cell death, is a highly regulated tumor suppressing process. However, functions and mechanisms of RNA-binding proteins in regulation of evasion of ferroptosis during lung cancer progression are still largely unknown. Here, we report that the RNA-binding protein RBMS1 participates in lung cancer development via mediating ferroptosis evasion. Through an shRNA-mediated systematic screen, we discovered that RBMS1 is a key ferroptosis regulator. Clinically, RBMS1 was elevated in lung cancer and its high expression was associated with reduced patient survival. Conversely, depletion of RBMS1 inhibited lung cancer progression both in vivo and in vitro. Mechanistically, RBMS1 interacted with the translation initiation factor eIF3d directly to bridge the 3'- and 5'-UTR of SLC7A11. RBMS1 ablation inhibited the translation of SLC7A11, reduced SLC7A11-mediated cystine uptake, and promoted ferroptosis. In a drug screen that targeted RBMS1, we further uncovered that nortriptyline hydrochloride decreased the level of RBMS1, thereby promoting ferroptosis. Importantly, RBMS1 depletion or inhibition by nortriptyline hydrochloride sensitized radioresistant lung cancer cells to radiotherapy. Our findings established RBMS1 as a translational regulator of ferroptosis and a prognostic factor with therapeutic potential and clinical value.
Keywords: Cell Biology; Lung cancer; Molecular biology; Oncology; Translation.
Figures





Similar articles
-
Suppressing circIDE/miR-19b-3p/RBMS1 axis exhibits promoting-tumour activity through upregulating GPX4 to diminish ferroptosis in hepatocellular carcinoma.Epigenetics. 2023 Dec;18(1):2192438. doi: 10.1080/15592294.2023.2192438. Epigenetics. 2023. PMID: 36989117 Free PMC article.
-
Inhibition of the RBMS1/PRNP axis improves ferroptosis resistance-mediated oxaliplatin chemoresistance in colorectal cancer.Mol Carcinog. 2024 Feb;63(2):224-237. doi: 10.1002/mc.23647. Epub 2023 Oct 20. Mol Carcinog. 2024. PMID: 37861356
-
RNA binding protein DAZAP1 promotes HCC progression and regulates ferroptosis by interacting with SLC7A11 mRNA.Exp Cell Res. 2021 Feb 1;399(1):112453. doi: 10.1016/j.yexcr.2020.112453. Epub 2020 Dec 29. Exp Cell Res. 2021. PMID: 33358859
-
Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy.Protein Cell. 2021 Aug;12(8):599-620. doi: 10.1007/s13238-020-00789-5. Epub 2020 Oct 1. Protein Cell. 2021. PMID: 33000412 Free PMC article. Review.
-
The amino acid transporter SLC7A11-mediated crosstalk implicated in cancer therapy and the tumor microenvironment.Biochem Pharmacol. 2022 Nov;205:115241. doi: 10.1016/j.bcp.2022.115241. Epub 2022 Sep 6. Biochem Pharmacol. 2022. PMID: 36084707 Review.
Cited by
-
Targeting ferroptosis regulators in lung cancer: Exploring natural products.Heliyon. 2024 Jul 2;10(14):e33934. doi: 10.1016/j.heliyon.2024.e33934. eCollection 2024 Jul 30. Heliyon. 2024. PMID: 39104501 Free PMC article. Review.
-
4EBP1-mediated SLC7A11 protein synthesis restrains ferroptosis triggered by MEK inhibitors in advanced ovarian cancer.JCI Insight. 2024 Jun 6;9(14):e177857. doi: 10.1172/jci.insight.177857. JCI Insight. 2024. PMID: 38842940 Free PMC article.
-
Emerging mechanisms of ferroptosis and its implications in lung cancer.Chin Med J (Engl). 2024 Apr 5;137(7):818-829. doi: 10.1097/CM9.0000000000003048. Epub 2024 Mar 18. Chin Med J (Engl). 2024. PMID: 38494343 Free PMC article. Review.
-
PRMT5-mediated FUBP1 methylation accelerates prostate cancer progression.J Clin Invest. 2024 Aug 15;134(18):e175023. doi: 10.1172/JCI175023. J Clin Invest. 2024. PMID: 39146021 Free PMC article.
-
Long-read subcellular fractionation and sequencing reveals the translational fate of full-length mRNA isoforms during neuronal differentiation.Genome Res. 2024 Nov 20;34(11):2000-2011. doi: 10.1101/gr.279170.124. Genome Res. 2024. PMID: 38839373 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

