A novel xenonucleic acid-mediated molecular clamping technology for early colorectal cancer screening
- PMID: 34610014
- PMCID: PMC8491914
- DOI: 10.1371/journal.pone.0244332
A novel xenonucleic acid-mediated molecular clamping technology for early colorectal cancer screening
Abstract
Background: Colorectal cancer (CRC) is one of the leading causes of cancer-related death. Early detection is critical to reduce CRC morbidity and mortality. In order to meet this need, we developed a molecular clamping assay called the ColoScape TM assay for early colorectal cancer diagnostics.
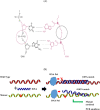
Methods: Nineteen mutations in four genes (APC, KRAS, BRAF and CTNNB1) associated with early events in CRC pathogenesis are targeted in the ColoScapeTM assay. Xenonucleic Acid (XNA)-mediated qPCR clamping technology was applied to minimize the wild-type background amplification in order to improve assay sensitivity of CRC mutation detection. The assay analytical performance was verified and validated, cfDNA and FFPE CRC patient samples were evaluated, and an ROC curve was applied to evaluate its performance.
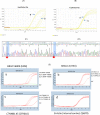
Results: The data showed that the assay analytical sensitivity was 0.5% Variant Allele Frequency, corresponding to ~7-8 copies of mutant DNA with 5 ng total DNA input per test. This assay is highly reproducible with intra-assay CV of <3% and inter-assay CV of <5%. We have investigated 380 clinical samples including plasma cfDNA and FFPE samples from patients with precancerous and different stages of CRC. The preliminary assay clinical specificity and sensitivity for CRC cfDNA were: 100% (95% CI, 80.3-97.5%) and 92.2% (95% CI, 94.7-100%), respectively, with AUC of 0.96; 96% specificity (95% CI, 77.6-99.7%) and 92% sensitivity (95% CI, 86.1-95.6%) with AUC of 0.94 for CRC FFPE; 95% specificity (95% CI, 82.5%-99.1%) and 62.5% sensitivity (95% CI, 35.8%-83.7%) with AUC of 0.79 for precancerous lesions cfDNA.
Conclusions: The XNA-mediated molecular clamping assay is a rapid, precise, and sensitive assay for the detection of precancerous lesions cfDNA and CRC cfDNA or FFPE samples.
Conflict of interest statement
The authors have read the journal’s policy and have the following competing interests: QS, LP, JD, MP, AZ, and MS are paid employees of Diacarta and WB is a Senior Adviser on the Medical Advisory Board of DiaCarta. There are no patents, products in development or marketed products associated with this research to declare. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures

Similar articles
-
Plasma mutation profile of precursor lesions and colorectal cancer using the Oncomine Colon cfDNA Assay.BMC Cancer. 2024 Dec 18;24(1):1547. doi: 10.1186/s12885-024-13287-2. BMC Cancer. 2024. PMID: 39695441 Free PMC article.
-
The potential of PIK3CA, KRAS, BRAF, and APC hotspot mutations as a non-invasive detection method for colorectal cancer.Mol Cell Probes. 2022 Jun;63:101807. doi: 10.1016/j.mcp.2022.101807. Epub 2022 Mar 13. Mol Cell Probes. 2022. PMID: 35296442
-
Analytical and clinical validation of a novel amplicon-based NGS assay for the evaluation of circulating tumor DNA in metastatic colorectal cancer patients.Clin Chem Lab Med. 2019 Sep 25;57(10):1501-1510. doi: 10.1515/cclm-2019-0142. Clin Chem Lab Med. 2019. PMID: 31339850
-
Circulating cell-free nucleic acids as biomarkers in colorectal cancer screening and diagnosis - an update.Expert Rev Mol Diagn. 2019 Jun;19(6):477-498. doi: 10.1080/14737159.2019.1613891. Epub 2019 May 21. Expert Rev Mol Diagn. 2019. PMID: 31046485 Review.
-
[Characteristics and diagnostic applications of circulating cell-free DNA in colorectal cancer].Orv Hetil. 2019 Jul;160(30):1167-1177. doi: 10.1556/650.2019.31486. Orv Hetil. 2019. PMID: 31327245 Review. Hungarian.
Cited by
-
Circulating Nucleic Acids in Colorectal Cancer: Diagnostic and Prognostic Value.Dis Markers. 2024 Feb 13;2024:9943412. doi: 10.1155/2024/9943412. eCollection 2024. Dis Markers. 2024. PMID: 38380073 Free PMC article. Review.
-
Staging of colorectal cancer using lipid biomarkers and machine learning.Metabolomics. 2023 Sep 20;19(10):84. doi: 10.1007/s11306-023-02049-z. Metabolomics. 2023. PMID: 37731020 Free PMC article.
-
A Highly Sensitive XNA-Based RT-qPCR Assay for the Identification of ALK, RET, and ROS1 Fusions in Lung Cancer.Diagnostics (Basel). 2024 Feb 24;14(5):488. doi: 10.3390/diagnostics14050488. Diagnostics (Basel). 2024. PMID: 38472960 Free PMC article.
-
A novel XNA-based Luminex assay to detect UBA1 somatic mutations associated with VEXAS syndrome.Pract Lab Med. 2024 Feb 24;39:e00380. doi: 10.1016/j.plabm.2024.e00380. eCollection 2024 Mar. Pract Lab Med. 2024. PMID: 38715663 Free PMC article.
References
-
- Kim et al. Colorectal Cancer Facts & Figures 2017–2019. the American Cancer Society Inc.1-36. https://www.cancer.org/research/cancer-facts-statistics/colorectal-cance...
-
- Levin B., Lieberman D.A., McFarland B., Smith R.A., Brooks D., Andrews K.S. et al.. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. CA Cancer J Clin. 2008; 58: 130–60. doi: 10.3322/CA.2007.0018 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

