BRCA1-BARD1 regulates transcription through modulating topoisomerase IIβ
- PMID: 34610268
- PMCID: PMC8492178
- DOI: 10.1098/rsob.210221
BRCA1-BARD1 regulates transcription through modulating topoisomerase IIβ
Abstract
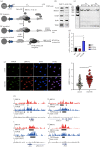
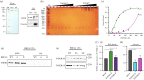
RNA polymerase II (Pol II)-dependent transcription in stimulus-inducible genes requires topoisomerase IIβ (TOP2B)-mediated DNA strand break and the activation of DNA damage response signalling in humans. Here, we report a novel function of the breast cancer 1 (BRCA1)-BRCA1-associated ring domain 1 (BARD1) complex in this process. We found that BRCA1 is phosphorylated at S1524 by the kinases ataxia-telangiectasia mutated and ATR during gene activation, and that this event is important for productive transcription. Our biochemical and genomic analyses showed that the BRCA1-BARD1 complex interacts with TOP2B in the EGR1 transcription start site and in a large number of protein-coding genes. Intriguingly, the BRCA1-BARD1 complex ubiquitinates TOP2B, which stabilizes TOP2B binding to DNA while BRCA1 phosphorylation at S1524 controls the TOP2B ubiquitination by the complex. Together, these findings suggest the novel function of the BRCA1-BARD1 complex in the regulation of TOP2B and Pol II-mediated gene expression.
Keywords: BRCA1-BARD1 complex; gene regulation; stimulus-inducible transcriptional activation; topoisomerase IIβ; transcription-coupled DNA break.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
