Overlapping genes in natural and engineered genomes
- PMID: 34611352
- PMCID: PMC8490965
- DOI: 10.1038/s41576-021-00417-w
Overlapping genes in natural and engineered genomes
Abstract
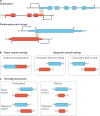
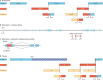
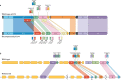
Modern genome-scale methods that identify new genes, such as proteogenomics and ribosome profiling, have revealed, to the surprise of many, that overlap in genes, open reading frames and even coding sequences is widespread and functionally integrated into prokaryotic, eukaryotic and viral genomes. In parallel, the constraints that overlapping regions place on genome sequences and their evolution can be harnessed in bioengineering to build more robust synthetic strains and constructs. With a focus on overlapping protein-coding and RNA-coding genes, this Review examines their discovery, topology and biogenesis in the context of their genome biology. We highlight exciting new uses for sequence overlap to control translation, compress synthetic genetic constructs, and protect against mutation.
© 2021. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Barrell BG, Air GM, Hutchison CA., 3rd Overlapping genes in bacteriophage phiX174. Nature. 1976;264:34–41. - PubMed
-
- Sanger F, et al. Nucleotide sequence of bacteriophage φX174 DNA. Nature. 1977;265:687. - PubMed
-
- Linney E, Hayashi M. Intragenic regulation of the synthesis of ΦX174 gene A proteins. Nature. 1974;249:345. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

