Expression and characterization of SARS-CoV-2 spike proteins
- PMID: 34611365
- PMCID: PMC9665560
- DOI: 10.1038/s41596-021-00623-0
Expression and characterization of SARS-CoV-2 spike proteins
Abstract
The severe acute respiratory syndrome coronavirus 2 spike protein is a critical component of coronavirus disease 2019 vaccines and diagnostics and is also a therapeutic target. However, the spike protein is difficult to produce recombinantly because it is a large trimeric class I fusion membrane protein that is metastable and heavily glycosylated. We recently developed a prefusion-stabilized spike variant, termed HexaPro for six stabilizing proline substitutions, that can be expressed with a yield of >30 mg/L in ExpiCHO cells. This protocol describes an optimized workflow for expressing and biophysically characterizing rationally engineered spike proteins in Freestyle 293 and ExpiCHO cell lines. Although we focus on HexaPro, this protocol has been used to purify over a hundred different spike variants in our laboratories. We also provide guidance on expression quality control, long-term storage, and uses in enzyme-linked immunosorbent assays. The entire protocol, from transfection to biophysical characterization, can be completed in 7 d by researchers with basic tissue cell culture and protein purification expertise.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
N.W. and J.S.M. are inventors on US patent application no. 62/412,703 (‘Prefusion Coronavirus Spike Proteins and Their Use’). D.W., N.W. and J.S.M. are inventors on US patent application no. 62/972,886 (‘2019-nCoV Vaccine’). C.-L.H., J.A.G., J.M.S., C.-W.C., A.M.D., K.J., H.-C.K., D.W., P.O.B., C.K.H., N.V.J., N.W., J.A.M., I.J.F. and J.S.M. are inventors on US patent application no. 63/032,502 (‘Engineered Coronavirus Spike (S) Protein and Methods of Use Thereof’).
Figures


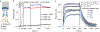

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

