Autophagy-dependent glutaminolysis drives superior IL21 production in HIV-1-specific CD4 T cells
- PMID: 34612140
- PMCID: PMC9225533
- DOI: 10.1080/15548627.2021.1972403
Autophagy-dependent glutaminolysis drives superior IL21 production in HIV-1-specific CD4 T cells
Abstract
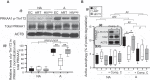
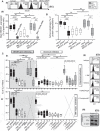
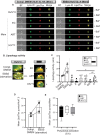
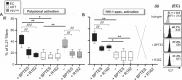
The maintenance of a strong IL21 production in memory CD4 T cells, especially in HIV-1-specific cells, represents a major correlate of natural immune protection against the virus. However, the molecular mechanisms underlying IL21 production during HIV-1 infection, which is only elevated among the naturally protected elite controllers (EC), are still unknown. We recently found out that lipophagy is a critical immune mediator that control an antiviral metabolic state following CD8A T cell receptor engagement, playing an important role in the natural control of HIV-1 infection. This led us to investigate whether the beneficial role of a strong macroautophagy/autophagy, could also be used to ensure effective IL21 production as well. Herein, we confirm that after both polyclonal and HIV-1-specific activation, memory CD4 T cells (Mem) from EC display enhanced activity of the autophagy-mediated proteolysis compared to ART. Our results indicate that the enhanced autophagy activity in EC was controlled by the energy-sensing PRKAA1 (protein kinase AMP-activated catalytic subunit alpha 1). We further confirmed the critical role of the autophagy-mediated proteolysis in the strong IL21 production in EC by using BECN1 gene silencing as well as protease, PRKAA1, and lysosomal inhibitors. Finally, we established that high autophagy-mediated proteolysis in EC fuels their cellular rates of mitochondrial respiration due to glutaminolysis. Our data confirm the critical role of autophagy in dictating the metabolic input, which is required not only to ensure protective cytotoxic CD8A T cell responses, but also to provide strong IL21 production among antiviral CD4 T cells.Abbreviations: AKG: alpha-ketoglutarate; ART: patients under antiretroviral therapy; ATG7: autophagy related 7; BaF: bafilomycin A1; BECN1: beclin 1; Chloro.: chloroquine; EC: elite controllers; EIF4EBP1: eukaryotic translation initiation factor 4E binding protein 1; FOXO3: forkhead box O3; GLS: glutaminase; GLUD1: glutamate dehydrogenase 1; HIVneg: HIV-1-uninfected control donors; IFNG/IFN-γ: interferon gamma; IL21: interleukin 21; MTOR: mechanistic target of rapamycin kinase; PBMC: peripheral blood mononuclear cells; PRKAA1: protein kinase AMP-activated catalytic subunit alpha 1; SQSTM1: sequestosome 1; TCA: tricarboxylic acid cycle; ULK1: unc-51 like autophagy activating kinase.
Keywords: Antiretroviral therapy; HIV-1; IL21; PRKAA1; autophagy-mediated proteolysis; elite controllers; glutaminolysis.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



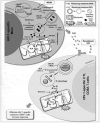


References
-
- Boucher CA, Bobkova MR, Geretti AM, et al. State of the art in HIV drug resistance: science and technology knowledge gap. AIDS Rev. 2018;20(1):27–42. - PubMed
-
- Dube MP, Sattler FR.. Inflammation and complications of HIV disease. J Infect Dis. 2010;201(12):1783–1785. - PubMed
-
- Warriner AH, Burkholder GA, Overton ET.. HIV-related metabolic comorbidities in the current ART era. Infect Dis Clin North Am. 2014;28(3):457–476. - PubMed
-
- Davenport MP, Khoury DS, Cromer D, et al. Functional cure of HIV: the scale of the challenge. Nat Rev Immunol. 2019;19(1):45–54. - PubMed
-
- Fonseca SG, Procopio FA, Goulet JP, et al. Unique features of memory T cells in HIV elite controllers: a systems biology perspective. Curr Opin HIV AIDS. 2011;6(3):188–196. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
