HIV-1 CA Inhibitors Are Antagonized by Inositol Phosphate Stabilization of the Viral Capsid in Cells
- PMID: 34613803
- PMCID: PMC8610598
- DOI: 10.1128/JVI.01445-21
HIV-1 CA Inhibitors Are Antagonized by Inositol Phosphate Stabilization of the Viral Capsid in Cells
Abstract
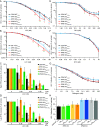
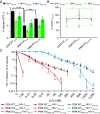
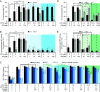
The HIV-1 capsid, composed of the CA protein, is the target of the novel antiretroviral drug lenacapavir (LCV). CA inhibitors block host factor binding and alter capsid stability to prevent nuclear entry and reverse transcription (RTN), respectively. Capsid stability is mediated in vitro by binding to the host cell metabolite inositol hexakisphosphate (IP6). IP6 depletion in target cells has little effect on HIV-1 infection. We hypothesized that capsid-altering concentrations of CA inhibitors might reveal an effect of IP6 depletion on HIV-1 infection in target cells. To test this, we studied the effects of IP6 depletion on inhibition of infection by the CA inhibitors PF74 and LCV. At low doses of either compound that affect HIV-1 nuclear entry, no effect of IP6 depletion on antiviral activity was observed. Increased antiviral activity was observed in IP6-depleted cells at inhibitor concentrations that affect capsid stability, correlating with increased RTN inhibition. Assays of uncoating and endogenous RTN of purified cores in vitro provided additional support. Our results show that inositol phosphates stabilize the HIV-1 capsid in target cells, thereby dampening the antiviral effects of capsid-targeting antiviral compounds. We propose that targeting of the IP6-binding site in conjunction with CA inhibitors will lead to robust antiretroviral therapy (ART). IMPORTANCE HIV-1 infection and subsequent depletion of CD4+ T cells result in AIDS. Antiretroviral therapy treatment of infected individuals prevents progression to AIDS. The HIV-1 capsid has recently become an ART target. Capsid inhibitors block HIV-1 infection at multiple steps, offering advantages over current ART. The cellular metabolite inositol hexakisphosphate (IP6) binds the HIV-1 capsid, stabilizing it in vitro. However, the function of this interaction in target cells is unclear. Our results imply that IP6 stabilizes the incoming HIV-1 capsid in cells, thus limiting the antiviral efficiency of capsid-destabilizing antivirals. We present a model of capsid inhibitor function and propose that targeting of the IP6-binding site in conjunction with capsid inhibitors currently in development will lead to more robust ART.
Keywords: CA; GS-6207; HIV; IP6; IPMK; IPPK; PF74; capsid; endogenous reverse transcription; lenacapavir.
Conflict of interest statement
We declare no competing interests.
Figures


References
-
- Rebensburg SV, Wei G, Larue RC, Lindenberger J, Francis AC, Annamalai AS, Morrison J, Shkriabai N, Huang SW, KewalRamani V, Poeschla EM, Melikyan GB, Kvaratskhelia M. 2021. Sec24C is an HIV-1 host dependency factor crucial for virus replication. Nat Microbiol 6:435–444. 10.1038/s41564-021-00868-1. - DOI - PMC - PubMed
-
- Achuthan V, Perreira JM, Sowd GA, Puray-Chavez M, McDougall WM, Paulucci-Holthauzen A, Wu X, Fadel HJ, Poeschla EM, Multani AS, Hughes SH, Sarafianos SG, Brass AL, Engelman AN. 2018. Capsid-CPSF6 interaction licenses nuclear HIV-1 trafficking to sites of viral DNA integration. Cell Host Microbe 24:392–404. 10.1016/j.chom.2018.08.002. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

