Targeting integrated epigenetic and metabolic pathways in lethal childhood PFA ependymomas
- PMID: 34613815
- PMCID: PMC8762577
- DOI: 10.1126/scitranslmed.abc0497
Targeting integrated epigenetic and metabolic pathways in lethal childhood PFA ependymomas
Abstract
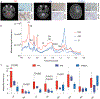
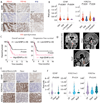
Childhood posterior fossa group A ependymomas (PFAs) have limited treatment options and bear dismal prognoses compared to group B ependymomas (PFBs). PFAs overexpress the oncohistone-like protein EZHIP (enhancer of Zeste homologs inhibitory protein), causing global reduction of repressive histone H3 lysine 27 trimethylation (H3K27me3), similar to the oncohistone H3K27M. Integrated metabolic analyses in patient-derived cells and tumors, single-cell RNA sequencing of tumors, and noninvasive metabolic imaging in patients demonstrated enhanced glycolysis and tricarboxylic acid (TCA) cycle metabolism in PFAs. Furthermore, high glycolytic gene expression in PFAs was associated with a poor outcome. PFAs demonstrated high EZHIP expression associated with poor prognosis and elevated activating mark histone H3 lysine 27 acetylation (H3K27ac). Genomic H3K27ac was enriched in PFAs at key glycolytic and TCA cycle–related genes including hexokinase-2 and pyruvate dehydrogenase. Similarly, mouse neuronal stem cells (NSCs) expressing wild-type EZHIP (EZHIP-WT) versus catalytically attenuated EZHIP-M406K demonstrated H3K27ac enrichment at hexokinase-2 and pyruvate dehydrogenase, accompanied by enhanced glycolysis and TCA cycle metabolism. AMPKα-2, a key component of the metabolic regulator AMP-activated protein kinase (AMPK), also showed H3K27ac enrichment in PFAs and EZHIP-WT NSCs. The AMPK activator metformin lowered EZHIP protein concentrations, increased H3K27me3, suppressed TCA cycle metabolism, and showed therapeutic efficacy in vitro and in vivo in patient-derived PFA xenografts in mice. Our data indicate that PFAs and EZHIP-WT–expressing NSCs are characterized by enhanced glycolysis and TCA cycle metabolism. Repurposing the antidiabetic drug metformin lowered pathogenic EZHIP, increased H3K27me3, and suppressed tumor growth, suggesting that targeting integrated metabolic/epigenetic pathways is a potential therapeutic strategy for treating childhood ependymomas.
Figures




References
-
- Pajtler KW, Witt H, Sill M, Jones DT, Hovestadt V, Kratochwil F, Wani K, Tatevossian R, Punchihewa C, Johann P, Reimand J, Warnatz HJ, Ryzhova M, Mack S, Ramaswamy V, Capper D, Schweizer L, Sieber L, Wittmann A, Huang Z, van Sluis P, Volckmann R, Koster J, Versteeg R, Fults D, Toledano H, Avigad S, Hoffman LM, Donson AM, Foreman N, Hewer E, Zitterbart K, Gilbert M, Armstrong TS, Gupta N, Allen JC, Karajannis MA, Zagzag D, Hasselblatt M, Kulozik AE, Witt O, Collins VP, von Hoff K, Rutkowski S, Pietsch T, Bader G, Yaspo ML, von Deimling A, Lichter P, Taylor MD, Gilbertson R, Ellison DW, Aldape K, Korshunov A, Kool M, Pfister SM, Molecular Classification of Ependymal Tumors across All CNS Compartments, Histopathological Grades, and Age Groups. Cancer Cell 27, 728–743 (2015). - PMC - PubMed
-
- Witt H, Mack SC, Ryzhova M, Bender S, Sill M, Isserlin R, Benner A, Hielscher T, Milde T, Remke M, Jones DT, Northcott PA, Garzia L, Bertrand KC, Wittmann A, Yao Y, Roberts SS, Massimi L, Van Meter T, Weiss WA, Gupta N, Grajkowska W, Lach B, Cho YJ, von Deimling A, Kulozik AE, Witt O, Bader GD, Hawkins CE, Tabori U, Guha A, Rutka JT, Lichter P, Korshunov A, Taylor MD, Pfister SM, Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell 20, 143–157 (2011). - PMC - PubMed
-
- Mack SC, Witt H, Piro RM, Gu L, Zuyderduyn S, Stutz AM, Wang X, Gallo M, Garzia L, Zayne K, Zhang X, Ramaswamy V, Jager N, Jones DT, Sill M, Pugh TJ, Ryzhova M, Wani KM, Shih DJ, Head R, Remke M, Bailey SD, Zichner T, Faria CC, Barszczyk M, Stark S, Seker-Cin H, Hutter S, Johann P, Bender S, Hovestadt V, Tzaridis T, Dubuc AM, Northcott PA, Peacock J, Bertrand KC, Agnihotri S, Cavalli FM, Clarke I, Nethery-Brokx K, Creasy CL, Verma SK, Koster J, Wu X, Yao Y, Milde T, Sin-Chan P, Zuccaro J, Lau L, Pereira S, Castelo-Branco P, Hirst M, Marra MA, Roberts SS, Fults D, Massimi L, Cho YJ, Van Meter T, Grajkowska W, Lach B, Kulozik AE, von Deimling A, Witt O, Scherer SW, Fan X, Muraszko KM, Kool M, Pomeroy SL, Gupta N, Phillips J, Huang A, Tabori U, Hawkins C, Malkin D, Kongkham PN, Weiss WA, Jabado N, Rutka JT, Bouffet E, Korbel JO, Lupien M, Aldape KD, Bader GD, Eils R, Lichter P, Dirks PB, Pfister SM, Korshunov A, Taylor MD, Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature 506, 445–450 (2014). - PMC - PubMed
-
- Merchant TE, Current Clinical Challenges in Childhood Ependymoma: A Focused Review. J. Clin. Oncol 35, 2364–2369 (2017). - PubMed
-
- Pajtler KW, Wen J, Sill M, Lin T, Orisme W, Tang B, Hubner JM, Ramaswamy V, Jia S, Dalton JD, Haupfear K, Rogers HA, Punchihewa C, Lee R, Easton J, Wu G, Ritzmann TA, Chapman R, Chavez L, Boop FA, Klimo P, Sabin ND, Ogg R, Mack SC, Freibaum BD, Kim HJ, Witt H, Jones DTW, Vo B, Gajjar A, Pounds S, Onar-Thomas A, Roussel MF, Zhang J, Taylor JP, Merchant TE, Grundy R, Tatevossian RG, Taylor MD, Pfister SM, Korshunov A, Kool M, Ellison DW, Molecular heterogeneity and CXorf67 alterations in posterior fossa group A (PFA) ependymomas. Acta Neuropathol. 136, 211–226 (2018). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K08 NS099427/NS/NINDS NIH HHS/United States
- F31 CA260935/CA/NCI NIH HHS/United States
- R01 CA204969/CA/NCI NIH HHS/United States
- R01 NS110572/NS/NINDS NIH HHS/United States
- T32 GM007863/GM/NIGMS NIH HHS/United States
- P50 CA097257/CA/NCI NIH HHS/United States
- R01 CA227622/CA/NCI NIH HHS/United States
- 22492/CRUK_/Cancer Research UK/United Kingdom
- T32 GM007315/GM/NIGMS NIH HHS/United States
- R01 CA222251/CA/NCI NIH HHS/United States
- 26398/CRUK_/Cancer Research UK/United Kingdom
- 28598/CRUK_/Cancer Research UK/United Kingdom

