Spatial organization of transcribing loci during early genome activation in Drosophila
- PMID: 34614388
- PMCID: PMC8612988
- DOI: 10.1016/j.cub.2021.09.027
Spatial organization of transcribing loci during early genome activation in Drosophila
Abstract
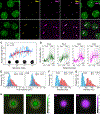
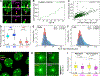
The early Drosophila embryo provides unique experimental advantages for addressing fundamental questions of gene regulation at multiple levels of organization, from individual gene loci to the entire genome. Using 1.5-h-old Drosophila embryos undergoing the first wave of genome activation,1 we detected ∼110 discrete "speckles" of RNA polymerase II (RNA Pol II) per nucleus, two of which were larger and localized to the histone locus bodies (HLBs).2,3 In the absence of the primary driver of Drosophila genome activation, the pioneer factor Zelda (Zld),1,4,5 70% fewer speckles were present; however, the HLBs tended to be larger than wild-type (WT) HLBs, indicating that RNA Pol II accumulates at the HLBs in the absence of robust early-gene transcription. We observed a uniform distribution of distances between active genes in the nuclei of both WT and zld mutant embryos, indicating that early co-regulated genes do not cluster into nuclear sub-domains. However, in instances whereby transcribing genes did come into close 3D proximity (within 400 nm), they were found to have distinct RNA Pol II speckles. In contrast to the emerging model whereby active genes are clustered to facilitate co-regulation and sharing of transcriptional resources, our data support an "individualist" model of gene control at early genome activation in Drosophila. This model is in contrast to a "collectivist" model, where active genes are spatially clustered and share transcriptional resources, motivating rigorous tests of both models in other experimental systems.
Keywords: RNA polymerase II; Zelda; genome activation; histone locus body; speckles.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
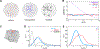

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

