B.1.617.2 enters and fuses lung cells with increased efficiency and evades antibodies induced by infection and vaccination
- PMID: 34614392
- PMCID: PMC8487035
- DOI: 10.1016/j.celrep.2021.109825
B.1.617.2 enters and fuses lung cells with increased efficiency and evades antibodies induced by infection and vaccination
Abstract
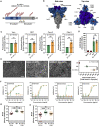
The Delta variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), B.1.617.2, emerged in India and has spread to over 80 countries. B.1.617.2 replaced B.1.1.7 as the dominant virus in the United Kingdom, resulting in a steep increase in new infections, and a similar development is expected for other countries. Effective countermeasures require information on susceptibility of B.1.617.2 to control by antibodies elicited by vaccines and used for coronavirus disease 2019 (COVID-19) therapy. We show, using pseudotyping, that B.1.617.2 evades control by antibodies induced upon infection and BNT162b2 vaccination, although to a lesser extent as compared to B.1.351. We find that B.1.617.2 is resistant against bamlanivimab, a monoclonal antibody with emergency use authorization for COVID-19 therapy. Finally, we show increased Calu-3 lung cell entry and enhanced cell-to-cell fusion of B.1.617.2, which may contribute to augmented transmissibility and pathogenicity of this variant. These results identify B.1.617.2 as an immune evasion variant with increased capacity to enter and fuse lung cells.
Keywords: B.1.617.2; COVID-19; SARS-CoV-2; antibody; neutralization; spike.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

References
-
- Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell. 2020;182:828–842.e16. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

