mRNA vaccination of naive and COVID-19-recovered individuals elicits potent memory B cells that recognize SARS-CoV-2 variants
- PMID: 34614412
- PMCID: PMC8452492
- DOI: 10.1016/j.immuni.2021.09.011
mRNA vaccination of naive and COVID-19-recovered individuals elicits potent memory B cells that recognize SARS-CoV-2 variants
Abstract
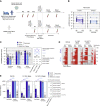
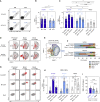
In addition to serum immunoglobulins, memory B cell (MBC) generation against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is another layer of immune protection, but the quality of MBC responses in naive and coronavirus disease 2019 (COVID-19)-recovered individuals after vaccination remains ill defined. We studied longitudinal cohorts of naive and disease-recovered individuals for up to 2 months after SARS-CoV-2 mRNA vaccination. We assessed the quality of the memory response by analysis of antibody repertoires, affinity, and neutralization against variants of concern (VOCs) using unbiased cultures of 2,452 MBCs. Upon boosting, the MBC pool of recovered individuals expanded selectively, matured further, and harbored potent neutralizers against VOCs. Although naive individuals had weaker neutralizing serum responses, half of their RBD-specific MBCs displayed high affinity toward multiple VOCs, including delta (B.1.617.2), and one-third retained neutralizing potency against beta (B.1.351). Our data suggest that an additional challenge in naive vaccinees could recall such affinity-matured MBCs and allow them to respond efficiently to VOCs.
Keywords: B-cell memory; BNT162b2 vaccine; COVID-19; RBD; affinity maturation; germinal center; neutralizing antibody; plasma cells; somatic hypermutation.
Copyright © 2021. Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests Outside of the submitted work, M. Mahévas. received research funds from GSK and personal fees from LFB and Amgen. J.-C.W. received consulting fees from Institut Mérieux. P.B. received consulting fees from Regeneron Pharmaceuticals. J.-M.P. received personal fees from Abbvie, Gilead, Merck, and Siemens Healthcare. F.R. is a member of the board of MELETIOS Therapeutics and of the Scientific Advisory Board of eureKARE.
Figures





References
-
- Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell. 2020;182:828–842.e16. - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

