An open-access volume electron microscopy atlas of whole cells and tissues
- PMID: 34616045
- PMCID: PMC9004664
- DOI: 10.1038/s41586-021-03992-4
An open-access volume electron microscopy atlas of whole cells and tissues
Erratum in
-
Publisher Correction: An open-access volume electron microscopy atlas of whole cells and tissues.Nature. 2021 Nov;599(7885):E5. doi: 10.1038/s41586-021-04132-8. Nature. 2021. PMID: 34732896 No abstract available.
Abstract
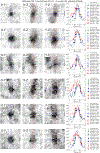
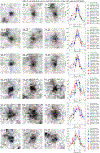
Understanding cellular architecture is essential for understanding biology. Electron microscopy (EM) uniquely visualizes cellular structures with nanometre resolution. However, traditional methods, such as thin-section EM or EM tomography, have limitations in that they visualize only a single slice or a relatively small volume of the cell, respectively. Focused ion beam-scanning electron microscopy (FIB-SEM) has demonstrated the ability to image small volumes of cellular samples with 4-nm isotropic voxels1. Owing to advances in the precision and stability of FIB milling, together with enhanced signal detection and faster SEM scanning, we have increased the volume that can be imaged with 4-nm voxels by two orders of magnitude. Here we present a volume EM atlas at such resolution comprising ten three-dimensional datasets for whole cells and tissues, including cancer cells, immune cells, mouse pancreatic islets and Drosophila neural tissues. These open access data (via OpenOrganelle2) represent the foundation of a field of high-resolution whole-cell volume EM and subsequent analyses, and we invite researchers to explore this atlas and pose questions.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
Portions of the technology described herein are covered by U.S. Patent 10,600,615 titled “Enhanced FIB-SEM systems for large-volume 3D imaging”, which was issued to C.S.X., K.J.H., and H.F.H., and assigned to Howard Hughes Medical Institute on March 24, 2020. The other authors declare no competing interests.
Figures











Comment in
-
Nanometre-scale imaging and AI reveal the interior of whole cells.Nature. 2021 Nov;599(7883):39-40. doi: 10.1038/d41586-021-02776-0. Nature. 2021. PMID: 34697481 No abstract available.
References
-
- Heinrich L et al. Automatic whole cell organelle segmentation in volumetric electron microscopy. bioRxiv 2020.11.14.382143 (2020). 10.1101/2020.11.13.382143 - DOI
-
- Lodish H et al. Molecular Cell Biology (W. H. Freeman, New York, 2016).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

