Key auxin response factor (ARF) genes constraining wheat tillering of mutant dmc
- PMID: 34616635
- PMCID: PMC8462377
- DOI: 10.7717/peerj.12221
Key auxin response factor (ARF) genes constraining wheat tillering of mutant dmc
Abstract
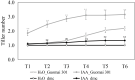
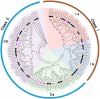
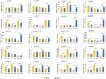
Tillering ability is a key agronomy trait for wheat (Triticum aestivum L.) production. Studies on a dwarf monoculm wheat mutant (dmc) showed that ARF11 played an important role in tillering of wheat. In this study, a total of 67 ARF family members were identified and clustered to two main classes with four subgroups based on their protein structures. The promoter regions of T. aestivum ARF (TaARF) genes contain a large number of cis-acting elements closely related to plant growth and development, and hormone response. The segmental duplication events occurred commonly and played a major role in the expansion of TaARFs. The gene collinearity degrees of the ARFs between wheat and other grasses, rice and maize, were significantly high. The evolution distances among TaARFs determine their expression profiles, such as homoeologous genes have similar expression profiles, like TaARF4-3A-1, TaARF4-3A-2 and their homoeologous genes. The expression profiles of TaARFs in various tissues or organs indicated TaARF3, TaARF4, TaARF9 and TaARF22 and their homoeologous genes played basic roles during wheat development. TaARF4, TaARF9, TaARF12, TaARF15, TaARF17, TaARF21, TaARF25 and their homoeologous genes probably played basic roles in tiller development. qRT-PCR analyses of 20 representative TaARF genes revealed that the abnormal expressions of TaARF11 and TaARF14 were major causes constraining the tillering of dmc. Indole-3-acetic acid (IAA) contents in dmc were significantly less than that in Guomai 301 at key tillering stages. Exogenous IAA application significantly promoted wheat tillering, and affected the transcriptions of TaARFs. These data suggested that TaARFs as well as IAA signaling were involved in controlling wheat tillering. This study provided valuable clues for functional characterization of ARFs in wheat.
Keywords: Auxin response factor; Expression profiles; IAA; Tillering; Wheat (Triticum aestivum L.).
©2021 Li et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures








References
-
- An J, Niu H, Ni Y, Jiang Y, Zheng Y, He R, Li J, Jiao Z, Zhang J, Li H, Li Q, Niu J. The miRNA-mRNA networks involving abnormal energy and hormone metabolisms restrict tillering in a wheat mutant dmc. International Journal of Molecular Sciences. 2019;20:4586. doi: 10.3390/ijms20184586. - DOI - PMC - PubMed
-
- Assuero SG, Lorenzo M, Ramírez NMP, Velázquez LM, Tognetti JA. Tillering promotion by paclobutrazol in wheat and its relationship with plant carbohydrate status. New Zealand Journal of Agricultural Research. 2012;55:347–358. doi: 10.1080/00288233.2012.706223. - DOI
LinkOut - more resources
Full Text Sources

