Smooth muscle cells in atherosclerosis: clones but not carbon copies
- PMID: 34617064
- PMCID: PMC8489213
- DOI: 10.1016/j.jvssci.2021.02.002
Smooth muscle cells in atherosclerosis: clones but not carbon copies
Erratum in
-
Correction.JVS Vasc Sci. 2024 Dec 25;5:100075. doi: 10.1016/j.jvssci.2022.04.003. eCollection 2024. JVS Vasc Sci. 2024. PMID: 40443980 Free PMC article.
Abstract
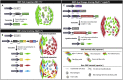
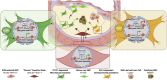
Our knowledge of the contribution of vascular smooth muscle cells (SMCs) to atherosclerosis has greatly advanced in the previous decade with the development of techniques allowing for the unambiguous identification and phenotypic characterization of SMC populations within the diseased vascular wall. By performing fate mapping or single-cell transcriptomics studies, or a combination of both, the field has made key observations: SMCs populate atherosclerotic lesions by the selective expansion and investment of a limited number of medial SMCs, which undergo profound and diverse modifications of their original phenotype and function. Thus, if SMCs residing within atherosclerotic lesions and contributing to the disease are clones, they are not carbon copies and can play atheroprotective or atheropromoting roles, depending on the nature of their phenotypic transitions. Tremendous progress has been made in identifying the transcriptional mechanisms biasing SMC fate. In the present review, we have summarized the recent advances in characterizing SMC investment and phenotypic diversity and the molecular mechanisms controlling SMC fate in atherosclerotic lesions. We have also discussed some of the remaining questions associated with these breakthrough observations. These questions include the underlying mechanisms regulating the phenomenon of SMC oligoclonal expansion; whether single-cell transcriptomics is reliable and sufficient to ascertain SMC functions and contributions during atherosclerosis development and progression; and how SMC clonality and phenotypic plasticity affects translational research and the therapeutic approaches developed to prevent atherosclerosis complications. Finally, we have discussed the complementary approaches the field should lean toward by combining single-cell phenotypic categorization and functional studies to understand further the complex SMC behavior and contribution in atherosclerosis.
Keywords: Cell differentiation; Cell plasticity; Coronary artery disease; Transcriptomics; Vascular cell; Vascular disease.
© 2021 by the Society for Vascular Surgery. Published by Elsevier Inc.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

