DL_FFLUX: A Parallel, Quantum Chemical Topology Force Field
- PMID: 34617748
- PMCID: PMC8582247
- DOI: 10.1021/acs.jctc.1c00595
DL_FFLUX: A Parallel, Quantum Chemical Topology Force Field
Abstract
DL_FFLUX is a force field based on quantum chemical topology that can perform molecular dynamics for flexible molecules endowed with polarizable atomic multipole moments (up to hexadecapole). Using the machine learning method kriging (aka Gaussian process regression), DL_FFLUX has access to atomic properties (energy, charge, dipole moment, etc.) with quantum mechanical accuracy. Newly optimized and parallelized using domain decomposition Message Passing Interface (MPI), DL_FFLUX is now able to deliver this rigorous methodology at scale while still in reasonable time frames. DL_FFLUX is delivered as an add-on to the widely distributed molecular dynamics code DL_POLY 4.08. For the systems studied here (103-105 atoms), DL_FFLUX is shown to add minimal computational cost to the standard DL_POLY package. In fact, the optimization of the electrostatics in DL_FFLUX means that, when high-rank multipole moments are enabled, DL_FFLUX is up to 1.25× faster than standard DL_POLY. The parallel DL_FFLUX preserves the quality of the scaling of MPI implementation in standard DL_POLY. For the first time, it is feasible to use the full capability of DL_FFLUX to study systems that are large enough to be of real-world interest. For example, a fully flexible, high-rank polarized (up to and including quadrupole moments) 1 ns simulation of a system of 10 125 atoms (3375 water molecules) takes 30 h (wall time) on 18 cores.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


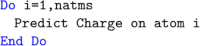
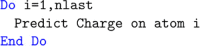


References
-
- Smith J. S.; Nebgen B. T.; Zubatyuk R.; Lubbers N.; Devereu C.; Barros K.; Tretiak S.; Isayev O.; Roitberg A. E. Approaching coupled cluster accuracy with a general-purpose neural network potential through transfer learning. Nat. Commun. 2019, 10, 290310.1038/s41467-019-10827-4. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

