Making sense of IL-6 signalling cues in pathophysiology
- PMID: 34618359
- PMCID: PMC9673051
- DOI: 10.1002/1873-3468.14201
Making sense of IL-6 signalling cues in pathophysiology
Abstract
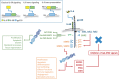
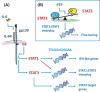
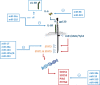
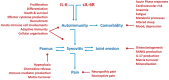
Unravelling the molecular mechanisms that account for functional pleiotropy is a major challenge for researchers in cytokine biology. Cytokine-receptor cross-reactivity and shared signalling pathways are considered primary drivers of cytokine pleiotropy. However, reports epitomized by studies of Jak-STAT cytokine signalling identify interesting biochemical and epigenetic determinants of transcription factor regulation that affect the delivery of signal-dependent cytokine responses. Here, a regulatory interplay between STAT transcription factors and their convergence to specific genomic enhancers support the fine-tuning of cytokine responses controlling host immunity, functional identity, and tissue homeostasis and repair. In this review, we provide an overview of the signalling networks that shape the way cells sense and interpret cytokine cues. With an emphasis on the biology of interleukin-6, we highlight the importance of these mechanisms to both physiological processes and pathophysiological outcomes.
Keywords: Jak-STAT signalling; STAT transcription factors; arthritis; cytokine receptors; cytokines; epigenetics; inflammation; interleukin; microRNA; pathophysiology.
© 2021 The Authors. FEBS Letters published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Figures
References
-
- Hunter CA and Jones SA (2015) IL‐6 as a keystone cytokine in health and disease. Nat Immunol 16, 448–457. - PubMed
-
- Garbers C, Heink S, Korn T and Rose‐John S (2018) Interleukin‐6: designing specific therapeutics for a complex cytokine. Nat Rev Drug Discov 17, 395–412. - PubMed
-
- Jones SA and Jenkins BJ (2018) Recent insights into targeting the IL‐6 cytokine family in inflammatory diseases and cancer. Nat Rev Immunol 18, 773–789. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

