Tumor-propagating side population cells are a dynamic subpopulation in undifferentiated pleomorphic sarcoma
- PMID: 34618689
- PMCID: PMC8663789
- DOI: 10.1172/jci.insight.148768
Tumor-propagating side population cells are a dynamic subpopulation in undifferentiated pleomorphic sarcoma
Abstract
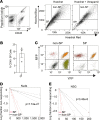
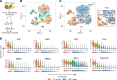
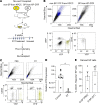
Sarcomas contain a subpopulation of tumor-propagating cells (TPCs) with enhanced tumor-initiating and self-renewal properties. However, it is unclear whether the TPC phenotype in sarcomas is stable or a dynamic cell state that can derive from non-TPCs. In this study, we utilized a mouse model of undifferentiated pleomorphic sarcoma (UPS) to trace the lineage relationship between sarcoma side population (SP) cells that are enriched for TPCs and non-SP cells. By cotransplanting SP and non-SP cells expressing different endogenous fluorescent reporters, we show that non-SP cells can give rise to SP cells with enhanced tumor-propagating potential in vivo. Lineage trajectory analysis using single-cell RNA sequencing from SP and non-SP cells supports the notion that non-SP cells can assume the SP cell phenotype de novo. To test the effect of eradicating SP cells on tumor growth and self-renewal, we generated mouse sarcomas in which the diphtheria toxin receptor is expressed in the SP cells and their progeny. Ablation of the SP population using diphtheria toxin did not impede tumor growth or self-renewal. Altogether, we show that the sarcoma SP represent a dynamic cell state and targeting TPCs alone is insufficient to eliminate tumor progression.
Keywords: Cancer; Mouse models; Oncology.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

