Metabolic Perturbation Associated With COVID-19 Disease Severity and SARS-CoV-2 Replication
- PMID: 34619366
- PMCID: PMC8490130
- DOI: 10.1016/j.mcpro.2021.100159
Metabolic Perturbation Associated With COVID-19 Disease Severity and SARS-CoV-2 Replication
Abstract
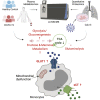
Viruses hijack host metabolic pathways for their replicative advantage. In this study, using patient-derived multiomics data and in vitro infection assays, we aimed to understand the role of key metabolic pathways that can regulate severe acute respiratory syndrome coronavirus-2 reproduction and their association with disease severity. We used multiomics platforms (targeted and untargeted proteomics and untargeted metabolomics) on patient samples and cell-line models along with immune phenotyping of metabolite transporters in patient blood cells to understand viral-induced metabolic modulations. We also modulated key metabolic pathways that were identified using multiomics data to regulate the viral reproduction in vitro. Coronavirus disease 2019 disease severity was characterized by increased plasma glucose and mannose levels. Immune phenotyping identified altered expression patterns of carbohydrate transporter, glucose transporter 1, in CD8+ T cells, intermediate and nonclassical monocytes, and amino acid transporter, xCT, in classical, intermediate, and nonclassical monocytes. In in vitro lung epithelial cell (Calu-3) infection model, we found that glycolysis and glutaminolysis are essential for virus replication, and blocking these metabolic pathways caused significant reduction in virus production. Taken together, we therefore hypothesized that severe acute respiratory syndrome coronavirus-2 utilizes and rewires pathways governing central carbon metabolism leading to the efflux of toxic metabolites and associated with disease severity. Thus, the host metabolic perturbation could be an attractive strategy to limit the viral replication and disease severity.
Keywords: COVID-19; mannose; metabolic transporters; metabolomics.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare no competing interests.
Figures





References
-
- Thomas T., Stefanoni D., Reisz J.A., Nemkov T., Bertolone L., Francis R.O., Hudson K.E., Zimring J.C., Hansen K.C., Hod E.A., Spitalnik S.L., D'Alessandro A. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight. 2020;5 - PMC - PubMed
-
- Su Y., Chen D., Yuan D., Lausted C., Choi J., Dai C.L., Voillet V., Duvvuri V.R., Scherler K., Troisch P., Baloni P., Qin G., Smith B., Kornilov S.A., Rostomily C., et al. Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell. 2020;183:1479–1495.e1420. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

