Anti-SARS-CoV-2 receptor-binding domain antibody evolution after mRNA vaccination
- PMID: 34619745
- PMCID: PMC8674133
- DOI: 10.1038/s41586-021-04060-7
Anti-SARS-CoV-2 receptor-binding domain antibody evolution after mRNA vaccination
Abstract
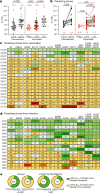
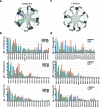
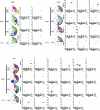
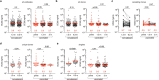
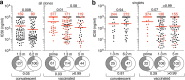
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection produces B cell responses that continue to evolve for at least a year. During that time, memory B cells express increasingly broad and potent antibodies that are resistant to mutations found in variants of concern1. As a result, vaccination of coronavirus disease 2019 (COVID-19) convalescent individuals with currently available mRNA vaccines produces high levels of plasma neutralizing activity against all variants tested1,2. Here we examine memory B cell evolution five months after vaccination with either Moderna (mRNA-1273) or Pfizer-BioNTech (BNT162b2) mRNA vaccine in a cohort of SARS-CoV-2-naive individuals. Between prime and boost, memory B cells produce antibodies that evolve increased neutralizing activity, but there is no further increase in potency or breadth thereafter. Instead, memory B cells that emerge five months after vaccination of naive individuals express antibodies that are similar to those that dominate the initial response. While individual memory antibodies selected over time by natural infection have greater potency and breadth than antibodies elicited by vaccination, the overall neutralizing potency of plasma is greater following vaccination. These results suggest that boosting vaccinated individuals with currently available mRNA vaccines will increase plasma neutralizing activity but may not produce antibodies with equivalent breadth to those obtained by vaccinating convalescent individuals.
© 2021. The Author(s).
Conflict of interest statement
The Rockefeller University has filed a provisional patent application in connection with this work on which M.C.N. is an inventor (US patent 63/021,387). The patent has been licensed by Rockefeller University to Bristol Meyers Squibb.
Figures

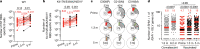






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

