Haplotype genomic prediction of phenotypic values based on chromosome distance and gene boundaries using low-coverage sequencing in Duroc pigs
- PMID: 34620094
- PMCID: PMC8496108
- DOI: 10.1186/s12711-021-00661-y
Haplotype genomic prediction of phenotypic values based on chromosome distance and gene boundaries using low-coverage sequencing in Duroc pigs
Abstract
Background: Genomic selection using single nucleotide polymorphism (SNP) markers has been widely used for genetic improvement of livestock, but most current methods of genomic selection are based on SNP models. In this study, we investigated the prediction accuracies of haplotype models based on fixed chromosome distances and gene boundaries compared to those of SNP models for genomic prediction of phenotypic values. We also examined the reasons for the successes and failures of haplotype genomic prediction.
Methods: We analyzed a swine population of 3195 Duroc boars with records on eight traits: body judging score (BJS), teat number (TN), age (AGW), loin muscle area (LMA), loin muscle depth (LMD) and back fat thickness (BF) at 100 kg live weight, and average daily gain (ADG) and feed conversion rate (FCR) from 30 to100 kg live weight. Ten-fold validation was used to evaluate the prediction accuracy of each SNP model and each multi-allelic haplotype model based on 488,124 autosomal SNPs from low-coverage sequencing. Haplotype blocks were defined using fixed chromosome distances or gene boundaries.
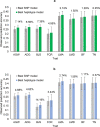
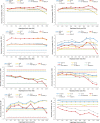
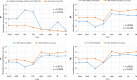
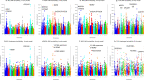
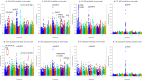
Results: Compared to the best SNP model, the accuracy of predicting phenotypic values using a haplotype model was greater by 7.4% for BJS, 7.1% for AGW, 6.6% for ADG, 4.9% for FCR, 2.7% for LMA, 1.9% for LMD, 1.4% for BF, and 0.3% for TN. The use of gene-based haplotype blocks resulted in the best prediction accuracy for LMA, LMD, and TN. Compared to estimates of SNP additive heritability, estimates of haplotype epistasis heritability were strongly correlated with the increase in prediction accuracy by haplotype models. The increase in prediction accuracy was largest for BJS, AGW, ADG, and FCR, which also had the largest estimates of haplotype epistasis heritability, 24.4% for BJS, 14.3% for AGW, 14.5% for ADG, and 17.7% for FCR. SNP and haplotype heritability profiles across the genome identified several genes with large genetic contributions to phenotypes: NUDT3 for LMA, LMD and BF, VRTN for TN, COL5A2 for BJS, BSND for ADG, and CARTPT for FCR.
Conclusions: Haplotype prediction models improved the accuracy for genomic prediction of phenotypes in Duroc pigs. For some traits, the best prediction accuracy was obtained with haplotypes defined using gene regions, which provides evidence that functional genomic information can improve the accuracy of haplotype genomic prediction for certain traits.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Boichard D, Guillaume F, Baur A, Croiseau P, Rossignol MN, Boscher MY, et al. Genomic selection in French dairy cattle. Anim Prod Sci. 2012;52:115–120. doi: 10.1071/AN11119. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

