Widespread bacterial diversity within the bacteriome of fungi
- PMID: 34621007
- PMCID: PMC8497576
- DOI: 10.1038/s42003-021-02693-y
Widespread bacterial diversity within the bacteriome of fungi
Abstract
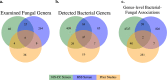
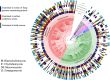
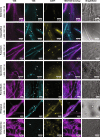
Knowledge of associations between fungal hosts and their bacterial associates has steadily grown in recent years as the number and diversity of examinations have increased, but current knowledge is predominantly limited to a small number of fungal taxa and bacterial partners. Here, we screened for potential bacterial associates in over 700 phylogenetically diverse fungal isolates, representing 366 genera, or a tenfold increase compared with previously examined fungal genera, including isolates from several previously unexplored phyla. Both a 16 S rDNA-based exploration of fungal isolates from four distinct culture collections spanning North America, South America and Europe, and a bioinformatic screen for bacterial-specific sequences within fungal genome sequencing projects, revealed that a surprisingly diverse array of bacterial associates are frequently found in otherwise axenic fungal cultures. We demonstrate that bacterial associations with diverse fungal hosts appear to be the rule, rather than the exception, and deserve increased consideration in microbiome studies and in examinations of microbial interactions.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

