Rapid Real-Time Polymerase Chain Reaction for Salmonella Serotyping Based on Novel Unique Gene Markers by Pangenome Analysis
- PMID: 34621261
- PMCID: PMC8491608
- DOI: 10.3389/fmicb.2021.750379
Rapid Real-Time Polymerase Chain Reaction for Salmonella Serotyping Based on Novel Unique Gene Markers by Pangenome Analysis
Abstract
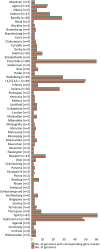
An accurate diagnostic method for Salmonella serovars is fundamental to preventing the spread of associated diseases. A diagnostic polymerase chain reaction (PCR)-based method has proven to be an effective tool for detecting pathogenic bacteria. However, the gene markers currently used in real-time PCR to detect Salmonella serovars have low specificity and are developed for only a few serovars. Therefore, in this study, we explored the novel unique gene markers for 60 serovars that share similar antigenic formulas and show high prevalence using pangenome analysis and developed a real-time PCR to detect them. Before exploring gene markers, the 535 Salmonella genomes were evaluated, and some genomes had serovars different from the designated serovar information. Based on these analyses, serovar-specific gene markers were explored. These markers were identified as genes present in all strains of target serovar genomes but absent in strains of other serovar genomes. Serovar-specific primer pairs were designed from the gene markers, and a real-time PCR method that can distinguish between 60 of the most common Salmonella serovars in a single 96-well plate assay was developed. As a result, real-time PCR showed 100% specificity for 199 Salmonella and 29 non-Salmonella strains. Subsequently, the method developed was applied successfully to both strains with identified serovars and an unknown strain, demonstrating that real-time PCR can accurately detect serovars of strains compared with traditional serotyping methods, such as antisera agglutination. Therefore, our method enables rapid and economical Salmonella serotyping compared with the traditional serotyping method.
Keywords: Salmonella; detection; gene marker; pangenome analysis; real-time PCR; serotyping.
Copyright © 2021 Yang, Kim, Kim, Kim, Baek, Ko, Kim, Yoon and Kim.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Abd-Elsalam K. A. (2003). Bioinformatic tools and guideline for PCR primer design. Afr. J. Biotechnol. 2 91–100. 10.5897/ajb2003.000-1019 - DOI
-
- Aboutalebian S., Ahmadikia K., Fakhim H., Chabavizadeh J., Okhovat A., Nikaeen M., et al. (2021). Direct Detection and Identification of the Most Common Bacteria and Fungi Causing Otitis Externa by a Stepwise Multiplex PCR. Front. Cell. Infect. Microbiol. 11:644060. 10.3389/fcimb.2021.644060 - DOI - PMC - PubMed
-
- Abubakar I., Irvine L., Aldus C. F., Wyatt G. M., Fordham R., Schelenz S., et al. (2007). A systematic review of the clinical, public health and cost-effectiveness of rapid diagnostic tests for the detection and identification of bacterial intestinal pathogens in faeces and food. Health Technol. Assess. 11 1–216. 10.3310/hta11360 - DOI - PubMed
-
- Broeders S., Huber I., Grohmann L., Berben G., Taverniers I., Mazzara M., et al. (2014). Guidelines for validation of qualitative real-time PCR methods. Trends Food Sci. Technol. 37 115–126. 10.1016/j.tifs.2014.03.008 - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

