Decoding Partner Specificity of Opioid Receptor Family
- PMID: 34621786
- PMCID: PMC8490921
- DOI: 10.3389/fmolb.2021.715215
Decoding Partner Specificity of Opioid Receptor Family
Abstract
This paper describes an exciting big data analysis compiled in a freely available database, which can be applied to characterize the coupling of different G-Protein coupled receptors (GPCRs) families with their intracellular partners. Opioid receptor (OR) family was used as case study in order to gain further insights into the physiological properties of these important drug targets, known to be associated with the opioid crisis, a huge socio-economic issue directly related to drug abuse. An extensive characterization of all members of the ORs family (μ (MOR), δ (DOR), κ (KOR), nociceptin (NOP)) and their corresponding binding partners (ARRs: Arr2, Arr3; G-protein: Gi1, Gi2, Gi3, Go, Gob, Gz, Gq, G11, G14, G15, G12, Gssh, Gslo) was carried out. A multi-step approach including models' construction (multiple sequence alignment, homology modeling), complex assembling (protein complex refinement with HADDOCK and complex equilibration), and protein-protein interface characterization (including both structural and dynamics analysis) were performed. Our database can be easily applied to several GPCR sub-families, to determine the key structural and dynamical determinants involved in GPCR coupling selectivity.
Keywords: G-protein; GPCRs; arrestin; database; functional signature; opioid receptor.
Copyright © 2021 Barreto, Baptista , Preto, Silvério, Melo and Moreira.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

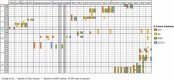
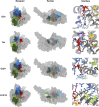
References
-
- Ballesteros J. A., Weinstein H. (1995). “[19] Integrated Methods for the Construction of Three-Dimensional Models and Computational Probing of Structure-Function Relations in G Protein-Coupled Receptors,” in Methods in Neurosciences. Receptor Molecular Biology. Editor Sealfon S. C. (Academic Press; ), 25, 366–428. 10.1016/S1043-9471(05)80049-7 - DOI
-
- Barreto C. A. V., Baptista S. J., Bueschbell B., Magalhães P., Preto A. J., Lemos A., et al. (2021) “Arrestin and G Protein Interactions with GPCRs: a Structural Perspective,” in GPCRs as Therapeutic Targets. 1st edn. (John Wiley and Sons Ltd)). (in press).
-
- Chang W., Cheng J., Allaire J., Sievert C., Schloerke B., Xie Y., et al. (2021). Shiny: Web Application Framework For R. R Package Version 1.6.0.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

