A reevaluation of the role of the ASIL trihelix transcription factors as repressors of the seed maturation program
- PMID: 34622120
- PMCID: PMC8483069
- DOI: 10.1002/pld3.345
A reevaluation of the role of the ASIL trihelix transcription factors as repressors of the seed maturation program
Abstract
Developmental transitions are typically tightly controlled at the transcriptional level. Two of these transitions involve the induction of the embryo maturation program midway through seed development and its repression during the vegetative phase of plant growth. Very little is known about the factors responsible for this regulation during early embryogenesis, and only a couple of transcription factors have been characterized as repressors during the postgerminative phase. Arabidopsis 6b-INTERACTING PROTEIN-LIKE1 (ASIL1), a trihelix transcription factor, has been proposed to repress maturation both embryonically and postembryonically. Preliminary data also suggested that its closest paralog, ASIL2, might play a role as well. We used a transcriptomic approach, coupled with phenotypical observations, to test the hypothesis that ASIL1 and ASIL2 redundantly turn off maturation during both phases of growth. Our results indicate that, contrary to what was previously published, neither of the ASIL genes plays a role in the regulation of maturation, at any point during plant development. Analyses of gene ontology (GO)-enriched terms and published transcriptomic datasets suggest that these genes might be involved in responses during the vegetative phase to certain biotic and abiotic stresses.
Keywords: Arabidopsis; embryo; embryonic maturation program; trihelix factor.
© 2021 The Authors. Plant Direct published by American Society of Plant Biologists and the Society for Experimental Biology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no conflicts of interests to declare.The Authors did not report any conflict of interest.
Figures
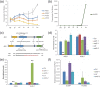
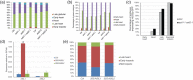




References
-
- Alonso, R., Oñate‐Sánchez, L., Weltmeier, F., Ehlert, A., Diaz, I., Dietrich, K., Vicente‐Carbajosa, J., & Dröge‐Laser, W. (2009). A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. The Plant Cell, 21(6), 1747–1761. 10.1105/tpc.108.062968 - DOI - PMC - PubMed
-
- Belmonte, M. F., Kirkbride, R. C., Stone, S. L., Pelletier, J. M., Bui, A. Q., Yeung, E. C., Hashimoto, M., Fei, J., Harada, C. M., Munoz, M. D., Le, B. H., Drews, G. N., Brady, S. M., Goldberg, R. B., & Harada, J. J. (2013). Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proceedings National Academy of Sciences USA, 110, E435–E444. 10.1073/pnas.1222061110 - DOI - PMC - PubMed

