Molecular predictors of response to pembrolizumab in thymic carcinoma
- PMID: 34622229
- PMCID: PMC8484507
- DOI: 10.1016/j.xcrm.2021.100392
Molecular predictors of response to pembrolizumab in thymic carcinoma
Abstract
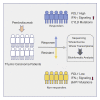
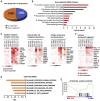
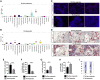
Thymic carcinoma is rare and has a poorer prognosis than thymomas. The treatment options are limited after failure of platinum-based chemotherapy. We previously performed a single-center phase II study of pembrolizumab in patients with advanced thymic carcinoma, showing a 22.5% response rate. Here, we characterize the genomic and transcriptomic profile of thymic carcinoma samples from 10 patients (5 non-responders versus 5 responders) in this cohort, with the main aim of identifying potential predictors of response to immunotherapy. We find that expression of PDL1 and alterations in genes or pathways that correlated with PD-L1 expression (CYLD and BAP1) could be potential predictors for response or resistance to immunotherapy in patients with advanced thymic carcinoma. Our study provides insights into potential predictive markers/pathways to select patients with thymic carcinoma for anti-PD-1 immunotherapy.
Trial registration: ClinicalTrials.gov NCT02364076.
Keywords: immune checkpoint inhibitors; predictors of response; thymic carcinoma; whole exome sequencing; whole-transcriptome sequencing.
© 2021 The Author(s).
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Greene M.A., Malias M.A. Aggressive multimodality treatment of invasive thymic carcinoma. J. Thorac. Cardiovasc. Surg. 2003;125:434–436. - PubMed
-
- Kondo K., Monden Y. Therapy for thymic epithelial tumors: a clinical study of 1,320 patients from Japan. Ann. Thorac. Surg. 2003;76:878–884, discussion 884–885. - PubMed
-
- Venuta F., Anile M., Diso D., Vitolo D., Rendina E.A., De Giacomo T., Francioni F., Coloni G.F. Thymoma and thymic carcinoma. Eur. J. Cardiothorac. Surg. 2010;37:13–25. - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

