Infant gut strain persistence is associated with maternal origin, phylogeny, and traits including surface adhesion and iron acquisition
- PMID: 34622230
- PMCID: PMC8484513
- DOI: 10.1016/j.xcrm.2021.100393
Infant gut strain persistence is associated with maternal origin, phylogeny, and traits including surface adhesion and iron acquisition
Abstract
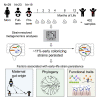
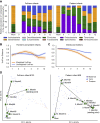
Gut microbiome succession affects infant development. However, it remains unclear what factors promote persistence of initial bacterial colonizers in the developing gut. Here, we perform strain-resolved analyses to compare gut colonization of preterm and full-term infants throughout the first year of life and evaluate associations between strain persistence and strain origin as well as genetic potential. Analysis of fecal metagenomes collected from 13 full-term and 9 preterm infants reveals that infants' initially distinct microbiomes converge by age 1 year. Approximately 11% of early colonizers, primarily Bacteroides and Bifidobacterium, persist during the first year of life, and those are more prevalent in full-term, compared with preterm infants. Examination of 17 mother-infant pairs reveals maternal gut strains are significantly more likely to persist in the infant gut than other strains. Enrichment in genes for surface adhesion, iron acquisition, and carbohydrate degradation may explain persistence of some strains through the first year of life.
Keywords: Infant gut microbiome; community ecology; early-life gut colonization; strain-resolved metagenomics.
© 2021.
Conflict of interest statement
J.F.B. is a cofounder of Metagenomi. The other authors declare no completing interests.
Figures





References
-
- Robertson R.C., Manges A.R., Finlay B.B., Prendergast A.J. The human microbiome and child growth—first 1000 days and beyond. Trends Microbiol. 2019;27:131–147. - PubMed
-
- Wang S., Ryan C.A., Boyaval P., Dempsey E.M., Ross R.P., Stanton C. Maternal vertical transmission affecting early-life microbiota development. Trends Microbiol. 2020;28:28–45. - PubMed
-
- Yassour M., Vatanen T., Siljander H., Hämäläinen A.-M., Härkönen T., Ryhänen S.J., Franzosa E.A., Vlamakis H., Huttenhower C., Gevers D., DIABIMMUNE Study Group Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med. 2016;8:343ra81. - PMC - PubMed

