Predicting endometrial cancer subtypes and molecular features from histopathology images using multi-resolution deep learning models
- PMID: 34622237
- PMCID: PMC8484685
- DOI: 10.1016/j.xcrm.2021.100400
Predicting endometrial cancer subtypes and molecular features from histopathology images using multi-resolution deep learning models
Abstract
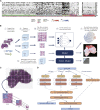
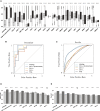
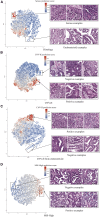
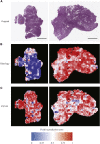
The determination of endometrial carcinoma histological subtypes, molecular subtypes, and mutation status is critical for the diagnostic process, and directly affects patients' prognosis and treatment. Sequencing, albeit slower and more expensive, can provide additional information on molecular subtypes and mutations that can be used to better select treatments. Here, we implement a customized multi-resolution deep convolutional neural network, Panoptes, that predicts not only the histological subtypes but also the molecular subtypes and 18 common gene mutations based on digitized H&E-stained pathological images. The model achieves high accuracy and generalizes well on independent datasets. Our results suggest that Panoptes, with further refinement, has the potential for clinical application to help pathologists determine molecular subtypes and mutations of endometrial carcinoma without sequencing.
Keywords: cancer genomics; cancer imaging; computational biology; computational pathology; deep learning; endometrial carcinoma.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Amant F., Moerman P., Neven P., Timmerman D., Van Limbergen E., Vergote I. Endometrial cancer. Lancet. 2005;366:491–505. - PubMed
-
- Morice P., Leary A., Creutzberg C., Abu-Rustum N., Darai E. Endometrial cancer. Lancet. 2016;387:1094–1108. - PubMed
-
- Burke W.M., Orr J., Leitao M., Salom E., Gehrig P., Olawaiye A.B., Brewer M., Boruta D., Villella J., Herzog T., Abu Shahin F., SGO Clinical Practice Endometrial Cancer Working Group. Society of Gynecologic Oncology Clinical Practice Committee Endometrial cancer: a review and current management strategies: part I. Gynecol. Oncol. 2014;134:385–392. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

