Ubiquitination of ATF6 by disease-associated RNF186 promotes the innate receptor-induced unfolded protein response
- PMID: 34623328
- PMCID: PMC8409591
- DOI: 10.1172/JCI145472
Ubiquitination of ATF6 by disease-associated RNF186 promotes the innate receptor-induced unfolded protein response
Abstract
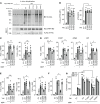
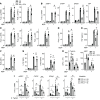
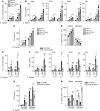
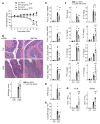
Properly balancing microbial responses by the innate immune system through pattern recognition receptors (PRRs) is critical for intestinal immune homeostasis. Ring finger protein 186 (RNF186) genetic variants are associated with inflammatory bowel disease (IBD). However, functions for the E3 ubiquitin ligase RNF186 are incompletely defined. We found that upon stimulation of the PRR nucleotide-binding oligomerization domain containing 2 (NOD2) in human macrophages, RNF186 localized to the ER, formed a complex with ER stress sensors, ubiquitinated the ER stress sensor activating transcription factor 6 (ATF6), and promoted the unfolded protein response (UPR). These events, in turn, led to downstream signaling, cytokine secretion, and antimicrobial pathway induction. Importantly, RNF186-mediated ubiquitination of K152 on ATF6 was required for these outcomes, highlighting a key role for ATF6 ubiquitination in PRR-initiated functions. Human macrophages transfected with the rare RNF186-A64T IBD risk variant and macrophages from common rs6426833 RNF186 IBD risk carriers demonstrated reduced NOD2-induced outcomes, which were restored by rescuing UPR signaling. Mice deficient in RNF186 or ATF6 demonstrated a reduced UPR in colonic tissues, increased weight loss, and less effective clearance of bacteria with dextran sodium sulfate-induced injury and upon oral challenge with Salmonella Typhimurium. Therefore, we identified that RNF186 was required for PRR-induced, UPR-associated signaling leading to key macrophage functions; defined that RNF186-mediated ubiquitination of ATF6 was essential for these functions; and elucidated how RNF186 IBD risk variants modulated these outcomes.
Keywords: Immunology; Innate immunity; Macrophages.
Conflict of interest statement
Figures









References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

