Inherited human c-Rel deficiency disrupts myeloid and lymphoid immunity to multiple infectious agents
- PMID: 34623332
- PMCID: PMC8409595
- DOI: 10.1172/JCI150143
Inherited human c-Rel deficiency disrupts myeloid and lymphoid immunity to multiple infectious agents
Abstract
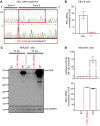
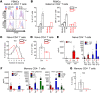
We studied a child with severe viral, bacterial, fungal, and parasitic diseases, who was homozygous for a loss-of-function mutation of REL, encoding c-Rel, which is selectively expressed in lymphoid and myeloid cells. The patient had low frequencies of NK, effector memory cells reexpressing CD45RA (Temra) CD8+ T cells, memory CD4+ T cells, including Th1 and Th1*, Tregs, and memory B cells, whereas the counts and proportions of other leukocyte subsets were normal. Functional deficits of myeloid cells included the abolition of IL-12 and IL-23 production by conventional DC1s (cDC1s) and monocytes, but not cDC2s. c-Rel was also required for induction of CD86 expression on, and thus antigen-presenting cell function of, cDCs. Functional deficits of lymphoid cells included reduced IL-2 production by naive T cells, correlating with low proliferation and survival rates and poor production of Th1, Th2, and Th17 cytokines by memory CD4+ T cells. In naive CD4+ T cells, c-Rel is dispensable for early IL2 induction but contributes to later phases of IL2 expression. The patient's naive B cells displayed impaired MYC and BCL2L1 induction, compromising B cell survival and proliferation and preventing their differentiation into Ig-secreting plasmablasts. Inherited c-Rel deficiency disrupts the development and function of multiple myeloid and lymphoid cells, compromising innate and adaptive immunity to multiple infectious agents.
Keywords: Genetic diseases; Genetics; Immunology.
Conflict of interest statement
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

