The N-terminal domain of SARS-CoV-2 nsp1 plays key roles in suppression of cellular gene expression and preservation of viral gene expression
- PMID: 34624207
- PMCID: PMC8481097
- DOI: 10.1016/j.celrep.2021.109841
The N-terminal domain of SARS-CoV-2 nsp1 plays key roles in suppression of cellular gene expression and preservation of viral gene expression
Abstract
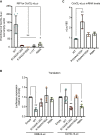
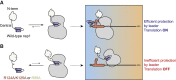
Nonstructural protein 1 (nsp1) is a coronavirus (CoV) virulence factor that restricts cellular gene expression by inhibiting translation through blocking the mRNA entry channel of the 40S ribosomal subunit and by promoting mRNA degradation. We perform a detailed structure-guided mutational analysis of severe acute respiratory syndrome (SARS)-CoV-2 nsp1, revealing insights into how it coordinates these activities against host but not viral mRNA. We find that residues in the N-terminal and central regions of nsp1 not involved in docking into the 40S mRNA entry channel nonetheless stabilize its association with the ribosome and mRNA, both enhancing its restriction of host gene expression and enabling mRNA containing the SARS-CoV-2 leader sequence to escape translational repression. These data support a model in which viral mRNA binding functionally alters the association of nsp1 with the ribosome, which has implications for drug targeting and understanding how engineered or emerging mutations in SARS-CoV-2 nsp1 could attenuate the virus.
Keywords: RNA; SARS; SARS-CoV-2; coronavirus; nsp1; nuclease; ribosome; translation.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests B.A.G. is a member of the Vaccine Advisory Board for Celsion Corporation.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

